Lassa mammarenavirus
Lassa mammarenavirus (LASV) is an arenavirus that causes Lassa hemorrhagic fever,[1] a type of viral hemorrhagic fever (VHF), in humans and other primates. Lassa mammarenavirus is an emerging virus and a select agent, requiring Biosafety Level 4-equivalent containment. It is endemic in West African countries, especially Sierra Leone, the Republic of Guinea, Nigeria, and Liberia, where the annual incidence of infection is between 300,000 and 500,000 cases, resulting in 5,000 deaths per year.[2]
| Lassa mammarenavirus | |
|---|---|
| TEM micrograph of Lassa mammarenavirus virions | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Ellioviricetes |
| Order: | Bunyavirales |
| Family: | Arenaviridae |
| Genus: | Mammarenavirus |
| Species: | Lassa mammarenavirus |
| Synonyms | |
| |
As of 2012 discoveries within the Mano River region of west Africa have expanded the endemic zone between the two known Lassa endemic regions, indicating that LASV is more widely distributed throughout the tropical wooded savannah ecozone in west Africa.[3] There are no approved vaccines against Lassa fever for use in humans.[4]
Discovery
In 1969, missionary nurse Laura Wine fell ill with a mysterious disease she contracted from an obstetrical patient in Lassa, a village in Borno State, Nigeria.[5][6][7] She was then transported to Jos, Nigeria where she died. Subsequently, two others became infected, one of whom was fifty-two-year-old nurse Lily Pinneo who had cared for Laura Wine.[8] Samples from Pinneo were sent to Yale University in New Haven where a new virus, that would later be known as Lassa mammarenavirus, was isolated for the first time by Jordi Casals and his team.[9][10] Casals contracted the fever, and nearly lost his life; one technician died from it.[9] By 1972, the multimammate rat, Mastomys natalensis, was found to be the main reservoir of the virus in West Africa, able to shed virus in its urine and feces without exhibiting visible symptoms.[11][12]
Virology
Structure and genome
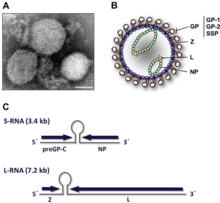
Lassa viruses[13][14] are enveloped, single-stranded, bisegmented, ambisense RNA viruses. Their genome[15] is contained in two RNA segments that code for two proteins each, one in each sense, for a total of four viral proteins.[16] The large segment encodes a small zinc finger protein (Z) that regulates transcription and replication,[17][18] and the RNA polymerase (L). The small segment encodes the nucleoprotein (NP) and the surface glycoprotein precursor (GP, also known as the viral spike), which is proteolytically cleaved into the envelope glycoproteins GP1 and GP2 that bind to the alpha-dystroglycan receptor and mediate host cell entry.[19]
Lassa fever causes hemorrhagic fever frequently shown by immunosuppression. Lassa mammarenavirus replicates very rapidly, and demonstrates temporal control in replication.[20] The first replication step is transcription of mRNA copies of the negative- or minus-sense genome. This ensures an adequate supply of viral proteins for subsequent steps of replication, as the NP and L proteins are translated from the mRNA. The positive- or plus-sense genome, then makes viral complementary RNA (vcRNA) copies of itself. The RNA copies are a template for producing negative-sense progeny, but mRNA is also synthesized from it. The mRNA synthesized from vcRNA are translated to make the GP and Z proteins. This temporal control allows the spike proteins to be produced last, and therefore, delay recognition by the host immune system.
Nucleotide studies of the genome have shown that Lassa has four lineages: three found in Nigeria and the fourth in Guinea, Liberia, and Sierra Leone. The Nigerian strains seem likely to have been ancestral to the others but additional work is required to confirm this.[21]
Receptors

Lassa mammarenavirus gains entry into the host cell by means of the cell-surface receptor the alpha-dystroglycan (alpha-DG),[19] a versatile receptor for proteins of the extracellular matrix. It shares this receptor with the prototypic Old World arenavirus lymphocytic choriomeningitis virus. Receptor recognition depends on a specific sugar modification of alpha-dystroglycan by a group of glycosyltransferases known as the LARGE proteins. Specific variants of the genes encoding these proteins appear to be under positive selection in West Africa where Lassa is endemic.[22] Alpha-dystroglycan is also used as a receptor by viruses of the New World clade C arenaviruses (Oliveros and Latino viruses). In contrast, the New World arenaviruses of clades A and B, which include the important viruses Machupo, Guanarito, Junin, and Sabia in addition to the non pathogenic Amapari virus, use the transferrin receptor 1. A small aliphatic amino acid at the GP1 glycoprotein amino acid position 260 is required for high-affinity binding to alpha-DG. In addition, GP1 amino acid position 259 also appears to be important, since all arenaviruses showing high-affinity alpha-DG binding possess a bulky aromatic amino acid (tyrosine or phenylalanine) at this position.
Unlike most enveloped viruses which use clathrin coated pits for cellular entry and bind to their receptors in a pH dependent fashion, Lassa and lymphocytic choriomeningitis virus instead use an endocytotic pathway independent of clathrin, caveolin, dynamin and actin. Once within the cell the viruses are rapidly delivered to endosomes via vesicular trafficking albeit one that is largely independent of the small GTPases Rab5 and Rab7. On contact with the endosome pH-dependent membrane fusion occurs mediated by the envelope glycoprotein, which at the lower pH of the endosome binds the lysosome protein LAMP1 which results in membrane fusion and escape from the endosome.
Life cycle
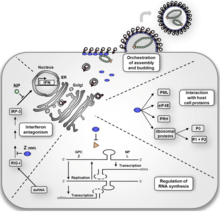
The life cycle of Lassa mammarenavirus is similar to the Old World arenaviruses. Lassa mammarenavirus enters the cell by the receptor-mediated endocytosis. Which endocytotic pathway is used is not known yet, but at least the cellular entry is sensitive to cholesterol depletion. It was reported that virus internalization is limited upon cholesterol depletion. The receptor used for cell entry is alpha-dystroglycan, a highly conserved and ubiquitously expressed cell surface receptor for extracellular matrix proteins. Dystroglycan, which is later cleaved into alpha-dystroglycan and beta-dystroglycan is originally expressed in most cells to mature tissues, and it provides molecular link between the ECM and the actin-based cytoskeleton.[23] After the virus enters the cell by alpha-dystroglycan mediated endocytosis, the low-pH environment triggers pH-dependent membrane fusion and releases RNP (viral ribonucleoprotein) complex into the cytoplasm. Viral RNA is unpacked, and replication and transcription initiate in the cytoplasm.[23] As replication starts, both S and L RNA genomes synthesize the antigenomic S and L RNAs, and from the antigenomic RNAs, genomic S and L RNA are synthesized. Both genomic and antigenomic RNAs are needed for transcription and translation. The S RNA encodes GP and NP (viral nucleocapsid protein) proteins, while L RNA encodes Z and L proteins. The L protein most likely represents the viral RNA-dependent RNA polymerase.[24] When the cell is infected by the virus, L polymerase is associated with the viral RNP and initiates the transcription of the genomic RNA. The 5’ and 3’ terminal 19 nt viral promoter regions of both RNA segments are necessary for recognition and binding of the viral polymerase. The primary transcription first transcribes mRNAs from the genomic S and L RNAs, which code NP and L proteins, respectively. Transcription terminates at the stem-loop (SL) structure within the intergenomic region. Arenaviruses use a cap snatching strategy to gain the cap structures from the cellular mRNAs, and it is mediated by the endonuclease activity of the L polymerase and the cap binding activity of NP. Antigenomic RNA transcribes viral genes GPC and Z, encoded in genomic orientation, from S and L segments respectively. The antigenomic RNA also serves as the template for the replication.[25] After translation of GPC, it is posttranslationally modified in the endoplasmic reticulum. GPC is cleaved into GP1 and GP2 at the later stage of the secretory pathway. It has been reported that the cellular protease SKI-1/S1P is responsible for this cleavage. The cleaved glycoproteins are incorporated into the virion envelope when the virus buds and release from the cell membrane.[24]
Pathogenesis
Lassa fever is mostly caused by the Lassa mammarenavirus. The symptoms include flu-like illness characterized by fever, general weakness, cough, sore throat, headache, and gastrointestinal manifestations. Hemorrhagic manifestations include vascular permeability.[25]
Upon entry, the Lassa mammarenavirus infects almost every tissue in the human body. It starts with the mucosa, intestine, lungs and urinary system, and then progresses to the vascular system.[5]
The main targets of the virus are antigen-presenting cells, mainly dendritic cells) and endothelial cells.[26][27][28] In 2012 it was reported how Lassa mammarenavirus nucleoprotein (NP) sabotages the host's innate immune system response. Generally, when a pathogen enters into a host, innate defense system recognizes the pathogen-associated molecular patterns (PAMP) and activates an immune response. One of the mechanisms detects double stranded RNA (dsRNA), which is only synthesized by negative-sense viruses. In the cytoplasm, dsRNA receptors, such as RIG-I (retinoic acid-inducible gene I) and MDA-5 (melanoma differentiation associated gene 5), detect dsRNAs and initiate signaling pathways that translocate IRF-3 (interferon regulatory factor 3) and other transcription factors to the nucleus. Translocated transcription factors activate expression of interferons 𝛂 and 𝛃, and these initiate adaptive immunity. NP encoded in Lassa mammarenavirus is essential in viral replication and transcription, but it also suppresses host innate IFN response by inhibiting translocation of IRF-3. NP of Lassa mammarenavirus is reported to have an exonuclease activity to only dsRNAs.[29] the NP dsRNA exonuclease activity counteracts IFN responses by digesting the PAMPs thus allowing the virus to evade host immune responses.[30]
References
- Frame JD, Baldwin JM, Gocke DJ, Troup JM (1 July 1970). "Lassa fever, a new virus disease of man from West Africa. I. Clinical description and pathological findings". Am. J. Trop. Med. Hyg. 19 (4): 670–6. doi:10.4269/ajtmh.1970.19.670. PMID 4246571.
- "Lassa Fever Fact Sheet".
- Sogoba, N.; Feldmann, H.; Safronetz, D. (14 November 2012). "Lassa Fever in West Africa: Evidence for an Expanded Region of Endemicity". Zoonoses & Public Health. 59 (59): 43–47. doi:10.1111/j.1863-2378.2012.01469.x. PMID 22958249.
- Yun, N; Walker, D (4 October 2012). "Pathogenesis of Lassa Fever". Viruses. 4 (10): 2031–2048. doi:10.3390/v4102031. PMC 3497040. PMID 23202452.
- Donaldson, Ross I. (2009). The Lassa Ward:One Man's Fight Against One of the World's Deadliest Diseases. St. Martin's Press. ISBN 0-312-37700-2. ISBN 978-0-312-37700-7.
- "Lassa Fever | CDC". www.cdc.gov. Retrieved 2016-09-23.
- Frame, J. D.; Baldwin, J. M.; Gocke, D. J.; Troup, J. M. (1970-07-01). "Lassa fever, a new virus disease of man from West Africa. I. Clinical description and pathological findings". The American Journal of Tropical Medicine and Hygiene. 19 (4): 670–676. doi:10.4269/ajtmh.1970.19.670. ISSN 0002-9637. PMID 4246571.
- Frame, J. D. (1992-05-01). "The story of Lassa fever. Part I: Discovering the disease". New York State Journal of Medicine. 92 (5): 199–202. ISSN 0028-7628. PMID 1614671.
- Prono, Luca (9 January 2008). Zhang, Yawei (ed.). Encyclopedia of Global Health. 1. SAGE. p. 354. ISBN 978-1-4129-4186-0. OCLC 775277696.
- Buckley, Sonja M.; Casals, Jordi; Downs, Wilbur G. (1970-07-11). "Isolation and Antigenic Characterization of Lassa Virus". Nature. 227 (5254): 174. Bibcode:1970Natur.227..174B. doi:10.1038/227174a0. PMID 5428406. S2CID 4211129.
- Fraser, D. W.; Campbell, C. C.; Monath, T. P.; Goff, P. A.; Gregg, M. B. (1974-11-01). "Lassa fever in the Eastern Province of Sierra Leone, 1970-1972. I. Epidemiologic studies". The American Journal of Tropical Medicine and Hygiene. 23 (6): 1131–1139. doi:10.4269/ajtmh.1974.23.1131. ISSN 0002-9637. PMID 4429182.
- Monath, T. P.; Maher, M.; Casals, J.; Kissling, R. E.; Cacciapuoti, A. (1974-11-01). "Lassa fever in the Eastern Province of Sierra Leone, 1970-1972. II. Clinical observations and virological studies on selected hospital cases". The American Journal of Tropical Medicine and Hygiene. 23 (6): 1140–1149. doi:10.4269/ajtmh.1974.23.1140. ISSN 0002-9637. PMID 4429183.
- Jamie Dyal and Ben Fohner Lassa virus Stanford University Humans and Viruses Class of 2005, n.d. accessed 9 May 2018
- Lashley, Felissa R., and Jerry D. Durham. Emerging Infectious Diseases: Trends and Issues. New York: Springer Pub., 2002. Print.
- Ridley, Matt. Genome: The Autobiography of a Species in 23 Chapters. New York: HarperCollins, 1999. Print.
- "Lassa virus RefSeq Genome".
- Cornu, T. I.; De La Torre, J. C. (2001). "RING Finger Z Protein of Lymphocytic Choriomeningitis Virus (LCMV) Inhibits Transcription and RNA Replication of an LCMV S-Segment Minigenome". Journal of Virology. 75 (19): 9415–9426. doi:10.1128/JVI.75.19.9415-9426.2001. PMC 114509. PMID 11533204.
- Djavani M, et al. (Sep 1997). "Completion of the Lassa fever virus sequence and identification of a RING finger open reading frame at the L RNA 5' End". Virology. 235 (2): 414–8. doi:10.1006/viro.1997.8722. PMID 9281522.
- Cao, W.; Henry, M. D.; Borrow, P.; Yamada, H.; Elder, J. H.; Ravkov, E. V.; Nichol, S. T.; Compans, R. W.; Campbell, K. P.; Oldstone, M. B. (1998). "Identification of -Dystroglycan as a Receptor for Lymphocytic Choriomeningitis Virus and Lassa Fever Virus". Science. 282 (5396): 2079–2081. Bibcode:1998Sci...282.2079C. doi:10.1126/science.282.5396.2079. PMID 9851928.
- Lashley, Felissa (2002). Emerging Infectious Diseases Trends and Issues. Springer Publishing Company.
- Bowen MD, Rollin PE, Ksiazek TG, et al. (August 2000). "Genetic Diversity among Lassa Virus Strains". J. Virol. 74 (15): 6992–7004. doi:10.1128/JVI.74.15.6992-7004.2000. PMC 112216. PMID 10888638.
- "Endemic: MedlinePlus Medical Encyclopedia".
- Rojek JM, Kunz S (April 2008). "Cell Entry by Human Pathogenic Arenaviruses". Cell Microbiol. 10 (4): 828–35. doi:10.1111/j.1462-5822.2007.01113.x. PMID 18182084.
- Drosten C, Kümmerer BM, Schmitz H, Günther S (January 2003). "Molecular Diagnostics of Viral Hemorrhagic Fevers". Antiviral Res. 57 (1–2): 61–87. doi:10.1016/s0166-3542(02)00201-2. PMID 12615304.
- Yun NE, Walker DH (October 2012). "Pathogenesis of Lassa Fever". Viruses. 4 (10): 2031–48. doi:10.3390/v4102031. PMC 3497040. PMID 23202452.
- Levene, M. I.; Gibson, N. A.; Fenton, A. C.; Papathoma, E.; Barnett, D. (1990). "The use of a calcium-channel blocker, nicardipine, for severely asphyxiated newborn infants". Developmental Medicine & Child Neurology. 32 (7): 567–574. doi:10.1111/j.1469-8749.1990.tb08540.x. PMID 2391009.
- Mahanty, S.; Hutchinson, K.; Agarwal, S.; McRae, M.; Rollin, P. E.; Pulendran, B. (2003). "Cutting edge: Impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses". Journal of Immunology. 170 (6): 2797–2801. doi:10.4049/jimmunol.170.6.2797. PMID 12626527.
- Baize, S.; Kaplon, J.; Faure, C.; Pannetier, D.; Georges-Courbot, M. C.; Deubel, V. (2004). "Lassa virus infection of human dendritic cells and macrophages is productive but fails to activate cells". Journal of Immunology. 172 (5): 2861–2869. doi:10.4049/jimmunol.172.5.2861. PMID 14978087.
- Hastie, Kathryn M.; King, Liam B.; Zandonatti, Michelle A.; Saphire, Erica Ollmann; Menéndez-Arias, Luis (Aug 2012). "Structural Basis for the dsRNA Specificity of the Lassa Virus NP Exonuclease". PLOS ONE. 7 (8): e44211. Bibcode:2012PLoSO...744211H. doi:10.1371/journal.pone.0044211. PMC 3429428. PMID 22937163.
- Hastie KM, Bale S, Kimberlin CR, Saphire EO (April 2012). "Hiding the evidence: two strategies for innate immune evasion by hemorrhagic fever viruses". Current Opinion in Virology. 2 (2): 151–6. doi:10.1016/j.coviro.2012.01.003. PMC 3758253. PMID 22482712.