p-Coumaric acid
p-Coumaric acid is a hydroxycinnamic acid, an organic compound that is a hydroxy derivative of cinnamic acid. There are three isomers of coumaric acid—o-coumaric acid, m-coumaric acid, and p-coumaric acid—that differ by the position of the hydroxy substitution of the phenyl group. p-Coumaric acid is the most abundant isomer of the three in nature. p-Coumaric acid exists in two forms trans-p-coumaric acid and cis-p-coumaric acid.
 | |
 | |
| Names | |
|---|---|
| Preferred IUPAC name
(2E)-3-(4-Hydroxyphenyl)prop-2-enoic acid | |
| Other names
(E)-3-(4-Hydroxyphenyl)-2-propenoic acid (E)-3-(4-Hydroxyphenyl)acrylic acid para-Coumaric acid 4-Hydroxycinnamic acid β-(4-Hydroxyphenyl)acrylic acid | |
| Identifiers | |
3D model (JSmol) |
|
| 2207383 | |
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| DrugBank | |
| ECHA InfoCard | 100.116.210 |
| EC Number |
|
| 2245630 | |
| KEGG | |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C9H8O3 | |
| Molar mass | 164.160 g·mol−1 |
| Melting point | 210 to 213 °C (410 to 415 °F; 483 to 486 K) |
| Hazards | |
| GHS pictograms | 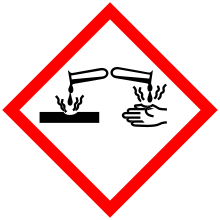   |
| GHS Signal word | Danger |
GHS hazard statements |
H301, H302, H311, H314, H315, H317, H319, H335 |
| P260, P261, P264, P270, P271, P272, P280, P301+310, P301+312, P301+330+331, P302+352, P303+361+353, P304+340, P305+351+338, P310, P312, P321, P322, P330, P332+313, P333+313, P337+313, P361, P362, P363 | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
It is a crystalline solid that is slightly soluble in water, but very soluble in ethanol and diethyl ether.
Natural occurrences
p-Coumaric acid can be found in Gnetum cleistostachyum.[1]
In food
p-Coumaric acid can be found in a wide variety of edible plants and fungi such as peanuts, navy beans, tomatoes, carrots, basil and garlic. It is found in wine and vinegar.[2] It is also found in barley grain.[3]
Derivatives
p-Coumaric acid glucoside can also be found in commercial breads containing flaxseed.[5]
Diesters of p-coumaric acid can be found in carnauba wax.
Metabolism
Biosynthesis
It is biosynthesized from cinnamic acid by the action of the P450-dependent enzyme 4-cinnamic acid hydroxylase (C4H).
It is also produced from L-tyrosine by the action of tyrosine ammonia lyase (TAL).

Biochemistry
p-Coumaric acid is the precursor of 4-ethylphenol produced by the yeast Brettanomyces in wine. The yeast converts this to 4-vinylphenol via the enzyme cinnamate decarboxylase.[6] 4-Vinylphenol is further reduced to 4-ethylphenol by the enzyme vinyl phenol reductase. Coumaric acid is sometimes added to microbiological media, enabling the positive identification of Brettanomyces by smell.
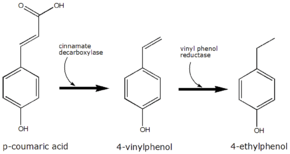 The conversion of p-coumaric acid to 4-ethyphenol by Brettanomyces
The conversion of p-coumaric acid to 4-ethyphenol by Brettanomyces
cis-p-Coumarate glucosyltransferase is an enzyme that uses uridine diphosphate glucose and cis-p-coumarate to produce 4′-O-β-D-glucosyl-cis-p-coumarate and uridine diphosphate (UDP). This enzyme belongs to the family of glycosyltransferases, specifically the hexosyltransferases.[7]
Phloretic acid is found in the rumen of sheep fed with dried grass and is produced by hydrogenation of the 2-propenoic side chain of p-coumaric acid.[8]
p-Coumaric acid serves as a photosensory chromophore when bound to a PYP domain.
As a cofactor of photoactive yellow proteins
p-Coumaric acid is the main enzyme cofactor of photoactive yellow proteins (PYP) which are a homologous group of proteins found in many eubacteria.[9] The importance of p-coumaric acid bound to PYP is due to its relative ease of forming stable crystals that diffract well for x-ray crystallography experiments, allowing biochemists and biophysicists to study the role of hydrogen bonding, molecular isomerization and photoactivity as biochemical phenomenon on a less complex scale than is present in other more complicated photosensitive proteins such as rhodopsin and cofactor retinal.[10][11][12][13]
Photochemical transitions
It was originally believed that due to light emissions resembling that of retinal bound rhodopsin, the photosensor molecule bound to PYP should resemble the structure of retinal bound rhodopsin, the photosensor molecule bound to PYP should resemble the structure of retinal.[14] Scientists were therefore amazed when the PYP Cys 69 was bound by a thiol ester linkage as the light sensitive prosthetic group p-coumaric acid.[9] During the photoreactive mechanism:[9][14]
- Light absorption yields the native protein to absorb a maximum wavelength of 446 nm, ε = 45500 M−1 cm−1.
- Within a nanosecond the absorbed maximum wavelength is shifted to 465 nm.
- Then on a sub-millisecond timescale is excited to a 355 nm state.
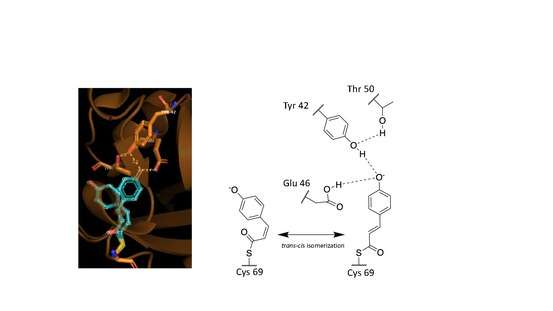
These observed phenomena are due to the trans–cis isomerization of the vinyl trans double bond in the p-coumaric acid.[9][13][12] Scientists noted by observing the crystal structure of p-coumaric acid bound by PYP that the hydroxyl group connected to the C4 carbon of the phenyl ring appeared to be deprotonated – effectively a phenolate functional group.[13][16] This was due to abnormally short hydrogen bonding lengths observed in the protein crystal structure.[15]
Role of hydrogen bonding
Hydrogen bonds in proteins such as PYP take part in interrelated networks, where at the center of p-coumaric acid's phenolate O4 atom, there is an oxyanion hole that is crucial for photosensory function.[12][17][18] Oxyanion holes exist in enzymes to stabilize transitions states of reaction intermediates, thus stabilizing the trans–cis isomerization of p-coumaric acid.[11][19] During the transition state it is believed that the p-coumaric acid phenolate O4 takes part in a hydrogen bond network between Glu46, Tyr42 and Thr50 of PYP.[19][11] These interactions are apart from the thiol ester linkage to Cys 69 keeping p-coumaric acid in the ligand binding cite.[9] Upon transitioning to the cis-isomeric form of p-coumaric acid the favorable hydrogen bonds are no longer in close interaction.
See also
- Coumarin
- Coumaroyl-Coenzyme A
- Ferulic acid
- Cinnamic acid
- Phenolic content in wine
- p-Coumaroylated anthocyanins
References
- Yao CS, Lin M, Liu X, Wang YH (April 2005). "Stilbene derivatives from Gnetum cleistostachyum". Journal of Asian Natural Products Research. 7 (2): 131–7. doi:10.1080/10286020310001625102. PMID 15621615.
- Gálvez MC, Barroso CG, Pérez-Bustamante JA (1994). "Analysis of polyphenolic compounds of different vinegar samples". Zeitschrift für Lebensmittel-Untersuchung und -Forschung. 199: 29–31. doi:10.1007/BF01192948.
- Quinde-Axtell Z, Baik BK (December 2006). "Phenolic compounds of barley grain and their implication in food product discoloration". Journal of Agricultural and Food Chemistry. 54 (26): 9978–84. doi:10.1021/jf060974w. PMID 17177530.
- Mao W, Schuler MA, Berenbaum MR (May 2013). "Honey constituents up-regulate detoxification and immunity genes in the western honey bee Apis mellifera". Proceedings of the National Academy of Sciences of the United States of America. 110 (22): 8842–6. Bibcode:2013PNAS..110.8842M. doi:10.1073/pnas.1303884110. PMC 3670375. PMID 23630255.
- Strandås C, Kamal-Eldin A, Andersson R, Åman P (October 2008). "Phenolic glucosides in bread containing flaxseed". Food Chemistry. 110 (4): 997–9. doi:10.1016/j.foodchem.2008.02.088. PMID 26047292.
- "Brettanomyces Monitoring by Analysis of 4-ethylphenol and 4-ethylguaiacol". etslabs.com. Archived from the original on 2008-02-19.
- Rasmussen S, Rudolph H (1997). "Isolation, purification and characterization of UDP-glucose: cis-p-coumaric acid-β-D-glucosyltransferase from sphagnum fallax". Phytochemistry. 46 (3): 449–453. doi:10.1016/S0031-9422(97)00337-3.
- Chesson A, Stewart CS, Wallace RJ (September 1982). "Influence of plant phenolic acids on growth and cellulolytic activity of rumen bacteria". Applied and Environmental Microbiology. 44 (3): 597–603. PMC 242064. PMID 16346090.
- Hoff WD, Düx P, Hård K, Devreese B, Nugteren-Roodzant IM, Crielaard W, Boelens R, Kaptein R, van Beeumen J, Hellingwerf KJ (November 1994). "Thiol ester-linked p-coumaric acid as a new photoactive prosthetic group in a protein with rhodopsin-like photochemistry". Biochemistry. 33 (47): 13959–62. doi:10.1021/bi00251a001. PMID 7947803.
- "PDB101: Molecule of the Month: Photoactive Yellow Protein". RCSB: PDB-101. Retrieved 2019-03-12.
- Pinney MM, Natarajan A, Yabukarski F, Sanchez DM, Liu F, Liang R, Doukov T, Schwans JP, Martinez TJ, Herschlag D (August 2018). "Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein". Journal of the American Chemical Society. 140 (31): 9827–9843. doi:10.1021/jacs.8b01596. PMID 29990421.
- Hellingwerf KJ (February 2000). "Key issues in the photochemistry and signalling-state formation of photosensor proteins". Journal of Photochemistry and Photobiology. B, Biology. 54 (2–3): 94–102. doi:10.1016/S1011-1344(00)00004-X. PMID 10836537.
- Premvardhan LL, Buda F, Van Der Horst MA, Lührs DC, Hellingwerf KJ, Van Grondelle R (2004-01-30). "Impact of Photon Absorption on the Electronic Properties of p -Coumaric Acid Derivatives of the Photoactive Yellow Protein Chromophore". The Journal of Physical Chemistry B. 108 (16): 5138–5148. doi:10.1021/jp037469b.
- Meyer TE, Yakali E, Cusanovich MA, Tollin G (January 1987). "Properties of a water-soluble, yellow protein isolated from a halophilic phototrophic bacterium that has photochemical activity analogous to sensory rhodopsin". Biochemistry. 26 (2): 418–23. doi:10.1021/bi00376a012. PMID 3828315.
- Genick UK, Borgstahl GE, Ng K, Ren Z, Pradervand C, Burke PM, Srajer V, Teng TY, Schildkamp W, McRee DE, Moffat K, Getzoff ED (March 1997). "Structure of a protein photocycle intermediate by millisecond time-resolved crystallography". Science. 275 (5305): 1471–5. doi:10.1126/science.275.5305.1471. PMID 9045611.
- Yamaguchi S, Kamikubo H, Kurihara K, Kuroki R, Niimura N, Shimizu N, Yamazaki Y, Kataoka M (January 2009). "Low-barrier hydrogen bond in photoactive yellow protein". Proceedings of the National Academy of Sciences of the United States of America. 106 (2): 440–4. Bibcode:2009PNAS..106..440Y. doi:10.1073/pnas.0811882106. PMC 2626721. PMID 19122140.
- Borgstahl GE, Williams DR, Getzoff ED (May 1995). "1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore". Biochemistry. 34 (19): 6278–87. doi:10.1021/bi00019a004. PMID 7756254.
- Stahl AD, Hospes M, Singhal K, van Stokkum I, van Grondelle R, Groot ML, Hellingwerf KJ (September 2011). "On the involvement of single-bond rotation in the primary photochemistry of photoactive yellow protein". Biophysical Journal. 101 (5): 1184–92. Bibcode:2011BpJ...101.1184S. doi:10.1016/j.bpj.2011.06.065. PMC 3164125. PMID 21889456.
- Herschlag D, Pinney MM (June 2018). "Hydrogen Bonds: Simple after All?". Biochemistry. 57 (24): 3338–3352. doi:10.1021/acs.biochem.8b00217. PMID 29678112.