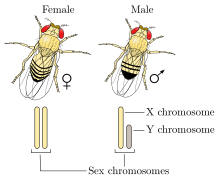
XY sex-determination system
The XY sex-determination system is the sex-determination system found in humans, most other mammals, some insects (Drosophila), some snakes, some fish (guppies), and some plants (Ginkgo tree). In this system, the sex of an individual is determined by a pair of sex chromosomes. Females typically have two of the same kind of sex chromosome (XX), and are called the homogametic sex. Males typically have two different kinds of sex chromosomes (XY), and are called the heterogametic sex.


| Part of a series on |
| Sex |
|---|
 |
| Biological terms |
| Sexual reproduction |
|
| Sexuality |
In humans, the presence of the Y chromosome is responsible for triggering male development; in the absence of the Y chromosome, the fetus will undergo female development. More specifically, it is the SRY gene located on the Y chromosome that is of importance to male differentiation. Variations to the sex gene karyotype could include rare disorders such as XX males (often due to translocation of the SRY gene to the X chromosome) or XY gonadal dysgenesis in people who are externally female (due to mutations in the SRY gene). In addition, other rare genetic variations such as Turners (XO) and Klinefelters (XXY) are seen as well.
The XY system contrasts in several ways with the ZW sex-determination system found in birds, some insects, many reptiles, and various other animals, in which the heterogametic sex is female. It had been thought for several decades that in all snakes sex was determined by the ZW system, but there had been observations of unexpected effects in the genetics of species in the families Boidae and Pythonidae; for example, parthenogenic reproduction produced only females rather than males, which is the opposite of what is to be expected in the ZW system. In the early years of the 21st century such observations prompted research that demonstrated that all pythons and boas so far investigated definitely have the XY system of sex determination.[1][2]
A temperature-dependent sex determination system is found in some reptiles.
Mechanisms
All animals have a set of DNA coding for genes present on chromosomes. In humans, most mammals, and some other species, two of the chromosomes, called the X chromosome and Y chromosome, code for sex. In these species, one or more genes are present on their Y chromosome that determine maleness. In this process, an X chromosome and a Y chromosome act to determine the sex of offspring, often due to genes located on the Y chromosome that code for maleness. Offspring have two sex chromosomes: an offspring with two X chromosomes will develop female characteristics, and an offspring with an X and a Y chromosome will develop male characteristics.
Humans
In humans, half of spermatozoa carry X chromosome and the other half Y chromosome.[3] A single gene (SRY) present on the Y chromosome acts as a signal to set the developmental pathway towards maleness. Presence of this gene starts off the process of virilization. This and other factors result in the sex differences in humans.[4] The cells in females, with two X chromosomes, undergo X-inactivation, in which one of the two X chromosomes is inactivated. The inactivated X chromosome remains within a cell as a Barr body.
Humans, as well as some other organisms, can have a rare chromosomal arrangement that is contrary to their phenotypic sex; for example, XX males or XY females (see androgen insensitivity syndrome). Additionally, an abnormal number of sex chromosomes (aneuploidy) may be present, such as Turner's syndrome, in which a single X chromosome is present, and Klinefelter's syndrome, in which two X chromosomes and a Y chromosome are present, XYY syndrome and XXYY syndrome.[4] Other less common chromosomal arrangements include: triple X syndrome, 48, XXXX, and 49, XXXXX.
Other animals
In most mammals, sex is determined by presence of the Y chromosome. Female is the default sex, due to the absence of the Y chromosome.[5] In the 1930s, Alfred Jost determined that the presence of testosterone was required for Wolffian duct development in the male rabbit.[6]
SRY is a sex-determining gene on the Y chromosome in the therians (placental mammals and marsupials).[7] Non-human mammals use several genes on the Y chromosome. Not all male-specific genes are located on the Y chromosome. Platypus, a monotreme, use five pairs of different XY chromosomes with six groups of male-linked genes, AMH being the master switch.[8] Other species (including most Drosophila species) use the presence of two X chromosomes to determine femaleness: one X chromosome gives putative maleness, but the presence of Y chromosome genes is required for normal male development.
Other systems
Birds and many insects have a similar system of sex determination (ZW sex-determination system), in which it is the females that are heterogametic (ZW), while males are homogametic (ZZ).[9]
Many insects of the order Hymenoptera instead have a haplo-diploid system, where the males are haploid (having just one chromosome of each type) while the females are diploid (with chromosomes appearing in pairs). Some other insects have the X0 sex-determination system, where just one chromosome type appears in pairs for the female but alone in the males, while all other chromosomes appear in pairs in both sexes.[10]
Influences
Genetic
In an interview for the Rediscovering Biology website,[11] researcher Eric Vilain described how the paradigm changed since the discovery of the SRY gene:
For a long time we thought that SRY would activate a cascade of male genes. It turns out that the sex determination pathway is probably more complicated and SRY may in fact inhibit some anti-male genes.
The idea is instead of having a simplistic mechanism by which you have pro-male genes going all the way to make a male, in fact there is a solid balance between pro-male genes and anti-male genes and if there is a little too much of anti-male genes, there may be a female born and if there is a little too much of pro-male genes then there will be a male born.
We [are] entering this new era in molecular biology of sex determination where it's a more subtle dosage of genes, some pro-males, some pro-females, some anti-males, some anti-females that all interplay with each other rather than a simple linear pathway of genes going one after the other, which makes it very fascinating but very complicated to study.
In mammals, including humans, the SRY gene is responsible with triggering the development of non-differentiated gonads into testes, rather than ovaries. However, there are cases in which testes can develop in the absence of an SRY gene (see sex reversal). In these cases, the SOX9 gene, involved in the development of testes, can induce their development without the aid of SRY. In the absence of SRY and SOX9, no testes can develop and the path is clear for the development of ovaries. Even so, the absence of the SRY gene or the silencing of the SOX9 gene are not enough to trigger sexual differentiation of a fetus in the female direction. A recent finding suggests that ovary development and maintenance is an active process,[12] regulated by the expression of a "pro-female" gene, FOXL2. In an interview[13] for the TimesOnline edition, study co-author Robin Lovell-Badge explained the significance of the discovery:
We take it for granted that we maintain the sex we are born with, including whether we have testes or ovaries. But this work shows that the activity of a single gene, FOXL2, is all that prevents adult ovary cells turning into cells found in testes.
Implications
Looking into the genetic determinants of human sex can have wide-ranging consequences. Scientists have been studying different sex determination systems in fruit flies and animal models to attempt an understanding of how the genetics of sexual differentiation can influence biological processes like reproduction, ageing[14] and disease.
Maternal
In humans and many other species of animals, the father determines the sex of the child. In the XY sex-determination system, the female-provided ovum contributes an X chromosome and the male-provided sperm contributes either an X chromosome or a Y chromosome, resulting in female (XX) or male (XY) offspring, respectively.
Hormone levels in the male parent affect the sex ratio of sperm in humans.[15] Maternal influences also impact which sperm are more likely to achieve conception.
Human ova, like those of other mammals, are covered with a thick translucent layer called the zona pellucida, which the sperm must penetrate to fertilize the egg. Once viewed simply as an impediment to fertilization, recent research indicates the zona pellucida may instead function as a sophisticated biological security system that chemically controls the entry of the sperm into the egg and protects the fertilized egg from additional sperm.[16]
Recent research indicates that human ova may produce a chemical which appears to attract sperm and influence their swimming motion. However, not all sperm are positively impacted; some appear to remain uninfluenced and some actually move away from the egg.[17]
Maternal influences may also be possible that affect sex determination in such a way as to produce fraternal twins equally weighted between one male and one female.[18]
The time at which insemination occurs during the estrus cycle has been found to affect the sex ratio of the offspring of humans, cattle, hamsters, and other mammals.[15] Hormonal and pH conditions within the female reproductive tract vary with time, and this affects the sex ratio of the sperm that reach the egg.[15]
Sex-specific mortality of embryos also occurs.[15]
History
Ancient ideas on sex determination
Aristotle believed incorrectly that the sex of an infant is determined by how much heat a man's sperm had during insemination. He wrote:
...the semen of the male differs from the corresponding secretion of the female in that it contains a principle within itself of such a kind as to set up movements also in the embryo and to concoct thoroughly the ultimate nourishment, whereas the secretion of the female contains material alone. If, then, the male element prevails it draws the female element into itself, but if it is prevailed over it changes into the opposite or is destroyed.
Aristotle claimed in error that the male principle was the driver behind sex determination,[19] such that if the male principle was insufficiently expressed during reproduction, the fetus would develop as a female.
20th century genetics
Nettie Stevens and Edmund Beecher Wilson are credited with independently discovering, in 1905, the chromosomal XY sex-determination system, i.e. the fact that males have XY sex chromosomes and females have XX sex chromosomes.[20][21][22]
The first clues to the existence of a factor that determines the development of testis in mammals came from experiments carried out by Alfred Jost,[23] who castrated embryonic rabbits in utero and noticed that they all developed as female.
In 1959, C. E. Ford and his team, in the wake of Jost's experiments, discovered[24] that the Y chromosome was needed for a fetus to develop as male when they examined patients with Turner's syndrome, who grew up as phenotypic females, and found them to be X0 (hemizygous for X and no Y). At the same time, Jacob & Strong described a case of a patient with Klinefelter syndrome (XXY),[25] which implicated the presence of a Y chromosome in development of maleness.[26]
All these observations lead to a consensus that a dominant gene that determines testis development (TDF) must exist on the human Y chromosome.[26] The search for this testis-determining factor (TDF) led a team of scientists[27] in 1990 to discover a region of the Y chromosome that is necessary for the male sex determination, which was named SRY (sex-determining region of the Y chromosome).[26]
See also
- Intersexuality for information on variations in human sexual forms
- Sexual differentiation (human)
- Secondary sex characteristic (human)
- Y-chromosomal Adam
- Sex Determination in Silene
References
- Gamble, Tony; Castoe, Todd A.; Nielsen, Stuart V.; Banks, Jaison L.; Card, Daren C.; Schield, Drew R.; Schuett, Gordon W.; Booth, Warren (2017). "The Discovery of XY Sex Chromosomes in a Boa and Python". Current Biology. 27 (14): 2148–2153.e4. doi:10.1016/j.cub.2017.06.010. PMID 28690112.
- Olena, Abby. Snake Sex Determination Dogma Overturned. The Scientist July 6, 2017
- "Five Facts about XX or XY". www.genderselectionauthority.com. March 4, 2014. Archived from the original on 2016-10-06.
- Fauci, Anthony S.; Braunwald, Eugene; Kasper, Dennis L.; Hauser, Stephen L.; Longo, Dan L.; Jameson, J. Larry; Loscalzo, Joseph (2008). Harrison's Principles of Internal Medicine (17th ed.). McGraw-Hill Medical. pp. 2339–2346. ISBN 978-0-07-147693-5.
- "Sex determination and differentiation" (PDF). Utrecht University - Department of Biology. Ultrecht, Netherlands. Archived from the original (PDF) on 27 November 2014. Retrieved 13 November 2014.
- Jost, A.; Price, D.; Edwards, R. G. (1970). "Hormonal Factors in the Sex Differentiation of the Mammalian Foetus [and Discussion]". Philosophical Transactions of the Royal Society B: Biological Sciences. 259 (828): 119–31. Bibcode:1970RSPTB.259..119J. doi:10.1098/rstb.1970.0052. JSTOR 2417046. PMID 4399057.
- Wallis MC, Waters PD, Graves JA (June 2008). "Sex determination in mammals - Before and after the evolution of SRY". Cell. Mol. Life Sci. 65 (20): 3182–95. doi:10.1007/s00018-008-8109-z. PMID 18581056.
- Cortez, Diego; Marin, Ray; Toledo-Flores, Deborah; Froidevaux, Laure; Liechti, Angélica; Waters, Paul D.; Grützner, Frank; Kaessmann, Henrik (24 April 2014). "Origins and functional evolution of Y chromosomes across mammals". Nature. 508 (7497): 488–493. Bibcode:2014Natur.508..488C. doi:10.1038/nature13151. PMID 24759410.
- Smith, Craig A.; Sinclair, Andrew H. (February 2004). "Sex determination: insights from the chicken". BioEssays. 26 (2): 120–132. doi:10.1002/bies.10400. ISSN 0265-9247. PMID 14745830.
- "5 Types of Sex Determination in Animals". genetics.knoji.com. Archived from the original on 5 February 2017. Retrieved 3 May 2018.
- Rediscovering Biology, Unit 11 - Biology of Sex and Gender, Expert interview transcripts, Link Archived 2010-08-23 at the Wayback Machine
- Uhlenhaut, N. Henriette; et al. (2009). "Somatic Sex Reprogramming of Adult Ovaries to Testes by FOXL2 Ablation". Cell. 139 (6): 1130–42. doi:10.1016/j.cell.2009.11.021. PMID 20005806.
- Scientists find single ‘on-off’ gene that can change gender traits Archived 2011-08-14 at the Wayback Machine, Hannah Devlin, The Times, December 11, 2009.
- Tower, John; Arbeitman, Michelle (2009). "The genetics of gender and life span". Journal of Biology. 8 (4): 38. doi:10.1186/jbiol141. PMC 2688912. PMID 19439039.
- Krackow, S. (1995). "Potential mechanisms for sex ratio adjustment in mammals and birds". Biological Reviews. 70 (2): 225–241. doi:10.1111/j.1469-185X.1995.tb01066.x. PMID 7605846.
- Suzanne Wymelenberg, Science and Babies, National Academy Press, 1990, page 17
- Richard E. Jones and Kristin H. Lopez, Human Reproductive Biology, Third Edition, Elsevier, 2006, page 238
- Familial recurrence of gender-balanced twins Archived October 2, 2015, at the Wayback Machine
- De Generatione Animalium, 766B 15‑17.
- Brush, Stephen G. (June 1978). "Nettie M. Stevens and the Discovery of Sex Determination by Chromosomes". Isis. 69 (2): 162–172. doi:10.1086/352001. JSTOR 230427. PMID 389882.
- "Nettie Maria Stevens – DNA from the Beginning". www.dnaftb.org. Archived from the original on 2012-10-01. Retrieved 2016-07-07.
- John L. Heilbron (ed.), The Oxford Companion to the History of Modern Science, Oxford University Press, 2003, "genetics".
- Jost A., Recherches sur la differenciation sexuelle de l’embryon de lapin, Archives d'anatomie microscopique et de morphologie experimentale, 36: 271 – 315, 1947.
- FORD CE, JONES KW, POLANI PE, DE ALMEIDA JC, BRIGGS JH (Apr 4, 1959). "A sex-chromosome anomaly in a case of gonadal dysgenesis (Turner's syndrome)". Lancet. 1 (7075): 711–3. doi:10.1016/S0140-6736(59)91893-8. PMID 13642858.
- JACOBS, PA; STRONG, JA (Jan 31, 1959). "A case of human intersexuality having a possible XXY sex-determining mechanism". Nature. 183 (4657): 302–3. Bibcode:1959Natur.183..302J. doi:10.1038/183302a0. PMID 13632697.
- Schoenwolf, Gary C. (2009). "Development of the Urogenital system". Larsen's human embryology (4th ed.). Philadelphia: Churchill Livingstone/Elsevier. pp. 307–9. ISBN 9780443068119.
- Sinclair, Andrew H.; et al. (19 July 1990). "A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif". Nature. 346 (6281): 240–244. Bibcode:1990Natur.346..240S. doi:10.1038/346240a0. PMID 1695712.
External links
- Sex Determination and Differentiation
- SRY: Sex determination from the National Center for Biotechnology Information
- Can Mammalian Mothers Control the Sex of their Offspring? (KQED Science article on San Diego Zoo research.)
- Maternal Diet and Other Factors Affecting Offspring Sex Ratio: A Review, published in Biology of Reproduction
- Sex Determination and the Maternal Dominance Hypothesis
- Sperm-Ovum Interactions at WikiGenes