Parvoviridae
Parvoviridae is a family of small, rugged, genetically-compact DNA viruses, known collectively as parvoviruses.[1][2] There are currently more than 100 species in the family, divided among 23 genera in three subfamilies.[3] Parvoviridae is the sole taxon in the order Quintoviricetes.
| Parvoviridae | |
|---|---|
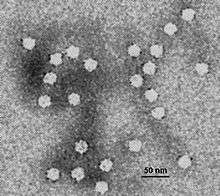 | |
| Electron micrograph of canine parvovirus | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Monodnaviria |
| Kingdom: | Shotokuvirae |
| Phylum: | Cossaviricota |
| Class: | Quintoviricetes |
| Order: | Piccovirales |
| Family: | Parvoviridae |
| Genera | |
Parvovirus B19 was the first pathogenic human parvovirus to be discovered and is best known for causing a childhood exanthem called "fifth disease" (erythema infectiosum), although it is also associated with other diseases including arthritis.
Parvoviruses can infect and may cause disease in many animals, from arthropods such as insects and shrimp, to echinoderms such as starfish, and to mammals including humans. Because most of these viruses require actively dividing cells to replicate, the type of tissue infected varies with the age of the animal. The gastrointestinal tract and lymphatic system can be affected at any age, leading to vomiting, diarrhea, and immunosuppression, but cerebellar hypoplasia is only seen in cats that were infected with feline parvovirus (FPV) in the womb or at less than two weeks of age, and disease of the myocardium is seen in puppies infected with canine parvovirus 2 (CPV2) between the ages of three and eight weeks.[4] Canine parvovirus causes a virulent and contagious disease in dogs. In cats, a parvovirus causes feline distemper.
Parvovirus can also be used specifically for members of one of the two Parvoviridae subfamilies: Parvovirinae, which infect vertebrate hosts, and Densovirinae, which infect invertebrate hosts, are more commonly referred to as densoviruses. In subfamily Densovirinae there are 5 genera and a total of 21 species.[2][3]
Parvoviruses are linear, nonsegmented, single-stranded DNA viruses, with an average genome size of 5-6 kilo base pairs (kbp). They are classified as group II viruses in the Baltimore classification of viruses. Parvoviruses are among the smallest viruses (hence the name, from Latin parvus meaning small) and are 23–28 nm in diameter.[5]
Structure
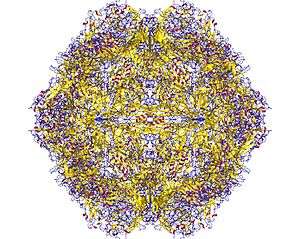
The viral capsid of a parvovirus is made up of 60 copies of two or more size variants of a single protein sequence, called VP1, VP2 etc., which form a resilient structure with T=1 icosahedral symmetry. These virions are typically resistant to dilute acids, bases, solvents, and temperatures up to 50°C (122°F). Parvoviruses do not have envelopes, thus are considered "naked" viruses. In addition, the shape of the virion is roughly spherical, with surface protrusions and canyons.[2]
Inside the capsid is a linear, single-stranded DNA genome in the size range 4–6 kbp, so the small genome of parvovirus can encode only a few proteins. At the 5’ and 3’ ends of this genome are short complementary sequences of roughly 120 to 550 nucleotides that form secondary structures as hairpins for example inverted terminal repeats (ITRs, which are two identical secondary structures at the termini) or unique sequences at the termini (two unique and different secondary structures are at each end of the DNA) and are essential for viral genome replication mechanism called rolling-hairpin replication.[2]
Parvovirus particles (virions) have a durable non-enveloped protein capsid ~20–30 nm in diameter that contains a single copy of the linear single-stranded ~ 5kb DNA genome, which terminates in small imperfect palindromes that fold into dynamic hairpin telomeres.[2][6][7][8] These terminal hairpins are hallmarks of the family, giving rise to the viral origins of DNA replication and mediating multiple steps in the viral life cycle including genome amplification, packaging, and the establishment of transcription complexes.[9][10] However, they are often refractory to detection by PCR amplification strategies since they tend to induce polymerase strand-switching.[11] Many parvoviruses are exceptionally resistant to inactivation, remaining infectious for months or years after release into the environment.[12][13]
Viruses in this family have small protein virions that exhibit T=1 icosahedral symmetry. Their capsid shells are assembled from 60 icosahedrally-ordered copies of a single core protein (VP) sequence, but some of these VP proteins also have N-terminal extensions that are not visible in X-ray structures.[6][7][8][9][10] Biochemical and serological studies indicate that these extensions become successively exposed at the particle surface during virus maturation and cell entry, where they contribute to virion stability and mediate specific steps in cell trafficking. Parvoviruses appear to be unique in encoding a broad spectrum phospholipase A2 (PLA2) activity, typically in the N-terminus of the longest (VP1) subset of their capsid proteins, which is deployed to mediate virion transfer across the lipid bilayer of host cells[14][15]
Size
These viruses have small genomes, encoding just two genes, and must rely on the synthetic machinery of their host cell for their own preferential replication. This means that many parvoviruses require host cells to enter S-phase before viral DNA replication can initiate, but they do not encode any gene products that can drive this transition. Parvoviruses overcome this problem in various ways: viruses in many genera simply wait within the cell for it to enter S-phase under its own cell cycle control, which means that they can only infect actively-dividing cell populations.
In contrast, the so-called adeno-associated viruses (AAVs) from genus Dependoparvovirus must wait until the cell is co-infected by a helper DNA virus, commonly an adenovirus or herpes virus, which does encode gene products that can drive the cell into S-phase, allowing AAV infection to initiate and out-compete the helper virus.[16]:137–146 A third strategy is used by human bocavirus 1 (HBoV1) from genus Bocaparvovirus, which appears to invoke a specific DNA-damage response in its host cell that ultimately supports viral DNA amplification and progeny virus production. [17]
Genome
The viral genome is 4–6 kilobases in length and terminates in imperfectly-palindromic hairpin sequences of ~120–500 nucleotides that exhibit genus-specific secondary structures, and can either be identical at the two ends of the genome (homotelomeric) or can differ in size, sequence and predicted secondary structure (heterotelomeric). Homotelomeric viruses package DNA strands of both senses (into separate capsids) whereas heterotelomeric viruses generally package predominantly negative-sense DNA (discussed in references 6 and 7). All parvoviruses encode two major gene complexes: the non-structural (or rep) gene that encodes the replication initiator protein (called NS1 or Rep), and the VP (or cap) gene, which encodes a nested set of ~2–6 size variants derived from the C-terminus of the single VP protein sequence. Members of the Parvovirinae also encode a few (1–4) small genus-specific ancillary proteins that are variably distributed throughout the genome, show little sequence homology to each other, and appear to serve an array of different functions in each genus (references 3–7). Viruses in most genera are mono-sense, meaning that both viral genes are transcribed in a single direction from open reading frames in the same (positive-sense) DNA strand, but members of one genus of homotelomeric invertebrate viruses (genus Ambidensovirus) show ambisense organization, with the NS and capsid proteins being transcribed in opposite directions from the 5’-ends of the two complementary DNA strands (see reference 1 and [18]).
The major non-structural protein, NS1, is a site- and strand-specific endonuclease belonging to the HuH protein superfamily,[19] and also carries a AAA+ SF3 helicase domain.[20] NS1 initiates and drives the viral “rolling hairpin” replication mechanism (RHR), which is a linear adaptation of the more-common “rolling-circle” replication strategy used by many small circular prokaryotic and viral replicons. RHR is a unidirectional mechanism that displaces a single, continuous DNA strand, which rapidly folds and refolds to generate a series of concatemeric duplex replication intermediates. Unit length genomes are then excised from these intermediates by the NS1 endonuclease (reviewed in references 5 and 6), and packaged 3’-to-5’ into preformed empty capsids driven by the SF3 helicase activity of NS1/Rep.[21]
Life cycle
Summary
Viral replication is nuclear. Entry into the host cell is achieved by attachment to host receptors, which mediate internalization via endocytosis. Capsids are metastable, undergoing a series of structural shifts during cell entry that sequentially expose peptides carrying PLA2 activity and trafficking signals . These signals ultimately mediate delivery of the intact virion into the cell nucleus, where genome uncoating allows the establishment of viral DNA replication and transcription complexes that rely predominantly upon the synthetic machinery of their host cell.[6][7][8][9][10][14][15] Replication follows the unidirectional strand displacement mechanism discussed above. Packaged virions from viruses in at least two genera (Protoparvovirus and Bocaparvovirus) have mechanisms that allow mature virions to be trafficked out of viable host cells prior to cell lysis, but members of most other genera are only released into the environment following death and lysis of the infected cell. Natural animal hosts for parvoviruses include a wide range of vertebrates, arthropods (insects and some crustacea) and echinoderms (sea stars). For viruses in the Parvovirinae transmission routes are commonly fecal-oral or respiratory.[22]
Steps of life cycle
Attachment and entry
A virion attaches to receptors on the surface of a potential host cell. In the case of B19 virus the host cell is a red blood cell precursor and one of the receptors is the blood group P antigen. The virion enters the cell by endocytosis and is released from the endosome into the cytoplasm, where it associates with microtubules and is transported to a nuclear pore. With a diameter of 18–26 nm, the parvovirus virion is small enough to pass through a nuclear pore, unlike the herpesvirus nucleocapsid, though there is evidence that the virion must undergo some structural changes before it can be transported into the nucleus. Nuclear localization signals have been found in the capsid proteins of some parvoviruses.[16]:141
Single-stranded DNA to double-stranded DNA
In the nucleus the single-stranded virus genome is converted to dsDNA by a cell DNA polymerase. The ends of the genome are double stranded as a result of base pairing, and at the 3' end the terminal nucleotides act as a primer to which the polymerase binds.[16]:141
Transcription and translation
The cell's RNA polymerase II transcribes the virus genes and cell transcription factors play key roles. The primary transcript(s) undergo various splicing events to produce several size classes of mRNA. The larger mRNAs encode the non-structural proteins and the smaller mRNAs encode the structural proteins. The non-structural proteins are phosphorylated and play roles in the control of gene expression and in DNA replication.[16]:142
DNA replication
After conversion of the ssDNA genome to dsDNA, the DNA is replicated by a mechanism called rolling-hairpin replication. This is a leading strand mechanism that is an adaptation of rolling-circle replication, and sets parvoviruses apart from dsDNA viruses, which replicate their genomes through leading and lagging strand synthesis. Procapsids are constructed from the structural proteins and each is filled by a copy of the virus genome, either a (+) DNA or a ( − ) DNA as appropriate. The helicase function of one of the non-structural proteins serves as a motor to drive the packaging of displaced single-strands into the capsid.[16]:142–3
Taxonomy
Group: ssDNA
The family is divided into three subfamilies: Parvovirinae, which infect vertebrates, Densovirinae, which infect invertebrates, and Hamaparvovirinae. Each subfamily has been subdivided into several genera.
Subfamily Densovirinae; genera:
- Aquabidensovirus
- Blattambidensovirus
- Hemiambidensovirus
- Iteradensovirus
- Miniambidensovirus
- Pefuambidensovirus
- Protoambidensovirus
- Scindoambidensovirus
Subfamily Hamaparvovirinae; genera:
- Brevihamaparvovirus
- Chaphamaparvovirus
- hepanhamaparvovirus
- Ichthamaparvovirus
- Penstylhamaparvovirus
Subfamily Parvovirinae:
- Genus Amdoparvovirus: type species: Carnivore amdoparvovirus 1. Genus includes 4 recognized species, infecting mink, fox, racoon dogs and skunk
- Genus Artiparvovirus
- Genus Aveparvovirus: type species: Galliform aveparvovirus 1. Genus includes a single species, infecting turkeys and chickens
- Genus Bocaparvovirus: type species: Ungulate bocaparvovirus 1. Genus includes 21 recognized species, infecting mammals from multiple orders, including primates
- Genus Copiparvovirus: type species: Ungulate copiparvovirus 1. Genus includes 2 recognized species, infecting pigs and cows
- Genus Dependoparvovirus: type species: Adeno-associated dependoparvovirus A. Genus includes 7 recognized species, infecting mammals, birds or reptiles
- Genus Erythroparvovirus: type species: Primate erythroparvovirus 1. Genus includes 6 recognized species, infecting mammals, specifically primates, chipmunk or cows
- Genus Loriparvovirus
- Genus Protoparvovirus: type species: Rodent protoparvovirus 1. Genus includes 11 recognized species, infecting mammals from multiple orders, including canines and primates
- Genus Tetraparvovirus: type species: Primate tetraparvovirus 1. Genus includes 6 recognized species, infecting primates, bats, pigs, cows and sheep
To date, very few viruses from the Densovirinae have been studied and sequenced, so the taxonomy may poorly reflect the true diversity of this subfamily. Currently, members of genus Ambidensovirus, Brevidensovirus and Iterodensovirus typically infect insects, while the hepan- and penstyldensoviruses infect decapod shrimp. However, recently two viruses have been isolated that segregate phylogenetically with members of genus Ambidensovirus but infect non-insect hosts. One of these, cherax quadricarinatus densovirus (CqDV), has been shown to infect and kill the freshwater crayfish Cherax quadricarinatus, subphylum Crustacea,[23] while the other, called sea-star-associated densovirus (SSaDV), has been shown to replicate in and kill sea-stars and perhaps some other members of phylum Echinodermata.[24]
Examples
Parvoviruses that infect vertebrate hosts make up the subfamily Parvovirinae, while those that infect invertabrates (currently only known to infect insects, crustacea, and echinoderms) make up the subfamily Densovirinae. Prior to 2014, the name parvovirus was also applied to a genus within subfamily Parvovirinae, but this genus name has been amended to Protoparvovirus to avoid confusion between taxonomic levels.[25][26]
Many mammalian species sustain infection by multiple parvoviruses. Parvoviruses tend to be specific about the species of animal they will infect, but this is a somewhat flexible characteristic. Thus, all isolates of canine parvovirus affect dogs, wolves, and foxes, but only some of them will infect cats.
Humans can be infected by viruses from five of the eight genera in the subfamily Parvovirinae: i) Bocaparvovirus (e.g. human bocavirus 1), ii) Dependoparvovirus (e.g. adeno-associated virus 2), iii) Erythroparvovirus (e.g. parvovirus B19), iv) Protoparvovirus (e.g. bufavirus 1a), and v) Tetraparvovirus (e.g. human parv4 G1). As of 2018, no known human viruses were in the remaining three recognized genera: vi) Amdoparvovirus (e.g. Aleutian mink disease virus), vii) Aveparvovirus (e.g. chicken parvovirus), and viii) Copiparvovirus (e.g. bovine parvovirus 2).[2]
Viruses infecting humans
Currently, viruses that infect humans are recognized in 5 genera: Bocaparvovirus (human bocavirus 1–4, HboV1–4), Dependoparvovirus (adeno-associated virus 1–5, AAV1–5), Erythroparvovirus (parvovirus B19, B19V), Protoparvovirus (bufavirus 1–3, BuV1–3; cutavirus, CuV) and Tetraparvovirus (human parvovirus 4 G1–3, PARV4 G1–3).
Dependoparvoviruses
Dependoparvoviruses, previously known as dependoviruses, require helper viruses (e.g. adenoviruses or herpesviruses) to replicate.[27] They are excellent vectors for experimental or clinical gene transduction and are now being used in patients for the therapeutic treatment of certain genetic diseases. The biggest advantage for such applications is that they are not known to cause any diseases.
Autonomous parvoviruses
Unlike dependoparvoviruse, autonomous parvoviruses do not require a helper virus. The autonomous parvovirus genome is a linear ssDNA with terminal palindromes, and several viruses have different sorts of palindromes at each end which are known to package the negative sense DNA strand for the most part. Most Autonomous Parvoviruses are known to encode only two NS structural proteins. Similar to RNA viruses, autonomous parvoviruses are highly effective when cells are in the S phase.
Autonomous Parvovirus Replication occurs through hairpins being formed by the palindrome at the 3′ end of the viral genome. This acts as a primer for the synthesis of complementary strands. Elongating strands then become covalently linked to hairpins at the 5' end of the template to create a linear duplex model that has covalent cross-links at the ends by DNA hairpins. Right end of the hairpin is nicked on the new strand by NS1. The genomic ssDNA is generated and packaged when the capsid is available. The RF structures create ssDNA but they are not linear duplex monomers. These Rf structures are potentially NS1/DNA complexes that are related to the structure of capsid.
Autonomous Parvoviruses have a very unique scheme of transcription. Several are known to utilize promoters at map regions 4 and 38 to create variable transcripts along with polyadenylation signals at the right end of the genome. Small introns exist between map positions 444 and 46, and large introns exist between map positions 10 and 39. All messengers in MVM have small introns spliced while alternative splice donors and acceptor sites can also be used. During genome replication, read-through of the internal poly S site occurs and the large transcript is translated into a capsid protein.
Protein synthesis is dependent on transcription. The earliest transcripts during the course of infection were the NS protein transcripts and they now are able to regulate gene expression. NS1 and NS2 are phosphorylated after translation, and NS1 is known to dimerize before nuclear localization. NS2 has been found to have profound impact on virus growth in a host-dependent manner.
Unclassified viruses
Since databases contain vast numbers of sequences that might be considered parvoviral in origin but are not real viruses, the Parvoviridae Study group cite the following criteria (references 1 and 13) that must be established before a new viral sequence can be considered for recognition in the family: "In order for an agent to be classified in the family Parvoviridae, it must be judged to be an authentic parvovirus on the basis of having been isolated and sequenced or, failing this, on the basis of having been sequenced in tissues, secretions, or excretions of unambiguous host origin, supported by evidence of its distribution in multiple individual hosts in a pattern that is compatible with dissemination by infection. The sequence must be in one piece, contain all the nonstructural (NS) and virus particle (VP) coding regions, and meet the size constraints and motif patterns typical of the family."
This means that partial coding sequences or sequences from a single host animal, with no evidence of virus exposure in the rest of the population, will not be considered sufficiently validated. Samples from feces are particularly problematic because they may be derived from food and so do not have an unequivocal host animal. Similarly, samples from aquatic animals that are also present in the surrounding environment are difficult to attribute until they can be shown to directly infect other members of the presumed host species. Despite these caveats, every year many new viruses are identified that will likely merit recognition, so that the published taxonomy always trails studies in the field.
For example, a candidate parvovirus has recently been isolated by sequencing a histocytic sarcoma in a slow loris (Nycticebus coucang).[28] The relationship between the virus and the sarcoma was not clear.
Additional genera
Possible new genera suggested in the literature include Chapparvovirus,[29] but these candidate viruses are predicted to have structurally distinct capsids. At present it is not clear if their host range is restricted to vertebrates, which could make them candidates for inclusion in subfamily Parvovirinae, or if they also infect invertebrate hosts. Consideration of the taxon awaits further investigation and ultimately a decision by the ICTV.
Diseases

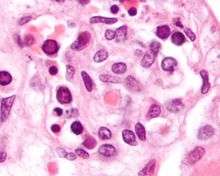
Parvovirus B19 which causes fifth disease in humans, is a member of species Primate erythroparvovirus 1 in the genus Erythroparvovirus. It infects red blood cell precursors and was the first parvovirus shown to cause human disease.[30] Some infections do not result in visible infection, while some manifest with visible effects, such as fifth disease (erythema infectiosum), which can give children a ‘slapped-cheek’ appearance.
Canine parvovirus is a member of species Carnivore protoparvovirus 1 in the genus Protoparvovirus. It causes a particularly deadly disease among young puppies, about 80% fatal, causing gastrointestinal tract damage and dehydration, as well as a cardiac syndrome in very young animals. It is spread by contact with an infected dog's feces. Symptoms include lethargy, severe diarrhea, fever, vomiting, loss of appetite, and dehydration.[31]
However, mouse parvovirus 1, a member of species Rodent protoparvovirus 1, causes no symptoms but can contaminate immunology experiments in biological research laboratories.
Porcine parvovirus, a member of species Ungulate Protoparvovirus 1, causes a reproductive disease in swine known as SMEDI, which stands for stillbirth, mummification, embryonic death, and infertility.
Like canine parvovirus, feline panleukopenia, now more commonly known as feline parvovirus, is also a member of species Carnivore protoparvovirus 1 in the genus Protoparvovirus. This virus is common in kittens and causes fever, low white blood cell count, diarrhea, and death. Infection of the cat fetus and kittens less than two weeks old causes cerebellar hypoplasia.
Mink enteritis virus, also a member of species Carnivore protoparvovirus 1, is similar in effect to feline parvovirus, except that it does not cause cerebellar hypoplasia. A different parvovirus causes aleutian disease in mink and other mustelids, characterized by lymphadenopathy, splenomegaly, glomerulonephritis, anemia, and death.
Canine and feline
Canine parvovirus is a mutant strain of feline parvovirus.[32][33][34] A very specific mutation is necessary for the virus to change species of infection. The mutation affects capsid proteins of feline parvovirus, giving it the ability to infect dogs.[35] Both forms of the virus are very similar, so once the mutation has occurred, canine parvovirus is still able to infect cats. The canine parvovirus has the tradeoff of gaining the ability to infect canine cells, while becoming less effective at infecting feline cells. Both feline parvovirus and canine parvovirus bind to and infect the transferrin receptors, but both have different sequences in the cells and animals. Infection by both feline parvovirus and canine parvovirus are relatively quick, but because of constant mutation of canine parvovirus, canine parvovirus has a slower infection time than feline parvovirus.[36] Studies of other strains of mutated canine parvovirus have revealed that changes in the viral capsid by just one protein can be fatal to the virus. Deleterious mutations have been noted to lead to inability to bind to transferrin receptors, bind to nonreceptive parts of the cell membrane, and identification of the virus by the host's antibody cells.[37]
Dogs, cats, and swine can be vaccinated against parvovirus. In pet dogs and cats, this is a normal vaccine commonly administered when they are young. The dog vaccine that protects against parvovirus is DHPP, and the cat vaccine is FPV.
Replication as disease vector
To enter host cells, parvoviruses typically bind to a sialic acid-bearing cell surface receptor and penetration into the cytoplasm is mediated by a phospholipase A2 activity carried on the amino-terminal peptide of the capsid VP1 polypeptide.[2] Once in the cytoplasm, the intact virus is translocated into the nucleus prior to uncoating. Transcription only initiates when the host cell enters S-phase under its own cell cycle control, when the cell's replication machinery converts the incoming single strand into a duplex transcription template, allowing synthesis of mRNAs encoding the nonstructural proteins, NS1 and NS2. The mRNAs are transported out of the nucleus into the cytoplasm, where the host ribosomes translate them into viral proteins. Viral DNA replication proceeds through a series of monomeric and concatemeric duplex intermediates by a unidirectional strand-displacement mechanism that is mediated by components of the host replication fork, aided and orchestrated by the viral NS1 polypeptide. NS1 also transactivates an internal transcriptional promoter that directs synthesis of the structural VP polypeptides. Once assembled capsids are available, replication shifts from synthesizing duplex DNA to displacement of progeny single strands, which are typically negative-sense and are packaged in a 3'-to-5' direction into formed particles within the nucleus. Mature virions may be released from infected cells prior to cell lysis, which promotes rapid transmission of the virus, but if this fails, then the virus is released at cell lysis.[2]
Unlike most other DNA viruses, parvoviruses are unable to activate DNA synthesis in host cells. Thus, for viral replication to take place, the infected cells must typically be nonquiescent (i.e. must be actively mitotic). Their inability to force host cells into S-phase means that parvoviruses are nontumorigenic. Indeed, they are commonly oncolytic, showing a strong tendency to replicate preferentially in cells with transformed phenotypes.[2]
Management and therapy
Currently, no vaccine exists to prevent infection by all parvoviruses, but recently, the virus's capsid proteins, which are noninfectious molecules, have been suggested acting as antigens for improving of vaccines. For pig vaccine, inactivated live, monovalent combined, most contain old PPV-1 strains to protect already positive sows. Vaccinate after 6 months, once or twice before mating, and repeat yearly.
Antivirals and human immunoglobulin-sourced treatments are usually for relief of symptoms. Using immunoglobulins is a logical solution for treatment as neutralizing antibodies because a majority of adults have been in danger from the parvoviruses, especially B19 virus.[38]
Use of HeLa cells in parvovirus testing
Testing for how feline parvovirus and canine parvovirus infect cells and what pathways are taken, scientists used cat cells, mouse cells, cat and mouse hybrid cells, mink cells, dog cells, human cells, and HeLa cells.[32] Both feline parvovirus and canine parvovirus enter their hosts, follow specific pathways, and infect at certain parts of cells before infecting major organs. Parvoviruses are specific viruses that are characterized in part by which receptors they attack.[33] Testing found that parvovirus infects carnivorous animals through the oropharyngeal pathway. Parvovirus infects the oropharyngeal cells by binding to transferrin receptors, a glycoprotein, on the plasma membrane.[34][35] The parvovirus plasmid is stored in a small non-enveloped capsid.[34][36] Once oropharyngeal cells become infected the virus spreads to dividing lymph cells and continues to work to the bone marrow and spread to target organs through blood.
Testing of HeLa cells and human cells to exposure of both feline parvovirus and canine parvovirus resulted in infections of the cells at human transferrin receptors.[32] When antibodies and parvovirus samples were added at the same time to human cells and HeLa cells, no infection was found to take place; both human cells and HeLa cells have transferrin receptors, but no evidence of humans contracting parvovirus was found.
Certain chromosomes in cells show more susceptibility to parvovirus than others. Testing of feline parvovirus on cat cells and cat mouse hybrid cells found cultures with cells having the highest concentrations of the C2 chromosome were the most highly infected cells.[32] Slight mutations of binding sites were found to slow down or completely stop the infection of the given parvovirus, whereas cells that are naturally missing the receptors or are mutants lacking them cannot be mutated.[35] Both feline parvovirus and canine parvovirus express plasticity during cellular infection.[36][37] Although transferrin receptors may be limited on cell surfaces, the parvovirus will find available transferrin receptors and will use different pathways to gain entry to the cell. Unlike plasma membrane infection plasticity, all strains of parvovirus show related routes to the cell nucleus.
Etymology
Viruses of the family Parvoviridae take their name from the Latin parvum, meaning "small" or "tiny". The subfamily Densovirinae has its name from the Latin denso, meaning thick or compact.[39]
History
Perhaps due to their extremely small size, the first parvoviruses were not discovered until the late 1950s.[40] Parvovirus B19, the first known parvovirus to cause disease in humans, was discovered in London by Australian virologist Yvonne Cossart in 1974. Cossart and her group were focused on hepatitis B and were processing blood samples when they discovered a number of "false positives" later identified as parvovirus B19. The virus is named for the patient code of one of the blood-bank samples involved in the discovery.[41]
See also
References
- Cotmore, SF; Agbandje-McKenna, M; Canuti, M; Chiorini, JA; Eis-Hubinger, A; Hughes, J; Mietzsch, M; Modha, S; Ogliastro, M; Pénzes, JJ; Pintel, DJ; Qiu, J; Soderlund-Venermo, M; Tattersall, P; Tijssen, P; and the ICTV Report Consortium (2019). "ICTV Virus Taxonomy Profile: Parvoviridae". Journal of General Virology. 100 (3): 367–368. doi:10.1099/jgv.0.001212. PMC 6537627. PMID 30672729.
- "ICTV 10th Report (2018) Parvoviridae".
- "Virus Taxonomy: 2019 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 8 May 2020.
- Fenner FJ, Gibbs EP, Murphy FA, Rott R, Studdert MJ, White DO (1993). Veterinary Virology (2nd ed.). Academic Press. ISBN 978-0-12-253056-2.
- Leppard K, Dimmock N, Easton A (2007). Introduction to Modern Virology. Blackwell. p. 450. ISBN 978-1-4051-3645-7.
- Berns KI, Parrish CR. 2013. Parvoviridae. In Fields Virology, ed. DM Knipe, P Howley. Philadelphia: Lippincott Williams & Wilkins. 6th ed
- Halder, S; Ng, R; Agbandje-McKenna, M (2012). "Parvoviruses: structure and infection". Future Virol. 7 (3): 253–7. doi:10.2217/fvl.12.12.
- Agbandje-McKenna, M; Kleinschmidt, J (2011). AAV capsid structure and cell interactions. Methods Mol. Biol. Methods in Molecular Biology. 807. pp. 47–92. doi:10.1007/978-1-61779-370-7_3. ISBN 978-1-61779-369-1. PMID 22034026.
- Cotmore, SF; Tattersall, P (2013). "Parvovirus diversity and DNA damage responses". Cold Spring Harb. Perspect. Biol. 5 (2): a01298. doi:10.1101/cshperspect.a012989. PMC 3552509. PMID 23293137.
- Cotmore, S.F.; Tattersall, P. (2014). "Parvoviruses: small does not mean simple". Annual Review of Virology. 1 (1): 517–537. doi:10.1146/annurev-virology-031413-085444. PMID 26958732.
- Huang, Q; Deng, X; Yan, Z; Cheng, F; Luo, Y; Shen, W; Lei-Butters, DC; Chen, AY; Li, Y; Tang, L; Söderlund-Venermo, M; Engelhardt, JF; Qiu, J (2012). "Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia". PLOS Pathog. 8 (8): e1002899. doi:10.1371/journal.ppat.1002899. PMC 3431310. PMID 22956907.
- Meriluoto, M; Hedman, L; Tanner, L; Simell, V; Mäkinen, M; et al. (2012). "Association of human bocavirus 1 infection with respiratory disease in childhood follow-up study, Finland". Emerg. Infect. Dis. 18 (2): 264–71. doi:10.3201/eid1802.111293. PMC 3310460. PMID 22305021.
- Truyen, U; Parrish, CR (2013). "Feline panleukopenia virus: its interesting evolution and current problems in immunoprophylaxis against a serious pathogen". Vet. Microbiol. 165 (1–2): 29–32. doi:10.1016/j.vetmic.2013.02.005. PMID 23561891.
- Zádori, Z; Szelei, J; Lacoste, M. C.; Li, Y; et al. (2001). "A viral phospholipase A2 (PLA2) is required for parvovirus infectivity". Dev. Cell. 1 (2): 291–302. doi:10.1016/s1534-5807(01)00031-4. PMID 11702787.
- Farr, GA; Zhang, LG; Tattersall, P (2005). "Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry". Proc. Natl. Acad. Sci. USA. 102 (47): 17148–53. Bibcode:2005PNAS..10217148F. doi:10.1073/pnas.0508477102. PMC 1288001. PMID 16284249.
- Carter J, Saunders V (2007). Virology: Principles and Applications. Wiley. ISBN 978-0-470-02386-0.
- Deng X, Yan Z, Cheng F, Engelhardt JF, Qiu J (2016). "Replication of an Autonomous Human Parvovirus in Non-dividing Human Airway Epithelium Is Facilitated through the DNA Damage and Repair Pathways". PLOS Pathogens. 12 (1): e1005399. doi:10.1371/journal.ppat.1005399. PMC 4713420. PMID 26765330.
- Cotmore, SF; Agbandje-McKenna, M; Chiorini, JA; Mukha, DV; Pintel, DJ; Qiu, J; Soderlund-Venermo, M; Tattersall, P; Tijssen, P; Gatherer, D; Davison, AJ (2014). "The family Parvoviridae". Arch. Virol. 159 (5): 1239–1247. doi:10.1007/s00705-013-1914-1. PMC 4013247. PMID 24212889.
- Hickman, AB; Ronning, DR; Perez, ZN; Kotin, RM; Dyda, F (2004). "The nuclease domain of adeno associated virus Rep coordinates replication initiation using two distinct DNA recognition interfaces". Mol. Cell. 13 (3): 403–14. doi:10.1016/s1097-2765(04)00023-1. PMID 14967147.
- James, JA; Aggarwal, AK; Linden, RM; Escalante, CR (2004). "Structure of adeno-associated virus type 2 Rep40-ADP complex: insight into nucleotide recognition and catalysis by superfamily 3 helicases". Proc. Natl. Acad. Sci. USA. 101 (34): 12455–60. Bibcode:2004PNAS..10112455J. doi:10.1073/pnas.0403454101. PMC 515083. PMID 15310852.
- King, JA; Dubielzig, R; Grimm, D; Kleinschmidt, JA (2001). "DNA helicase–mediated packaging of adeno-associated virus type 2 genomes into preformed capsids". EMBO J. 20 (12): 3282–91. doi:10.1093/emboj/20.12.3282. PMC 150213. PMID 11406604.
- "Viral Zone". ExPASy. Retrieved 15 June 2015.
- Bochow, S; Condon, K; Elliman, J; Owens, L (2015). "First complete genome of an Ambidensovirus; Cherax quadricarinatus densovirus, from freshwater crayfish Cherax quadricarinatus". Marine Genomics. 24: 305–312. doi:10.1016/j.margen.2015.07.009. PMID 26268797.
- Hewson, I; Button, JB; Gudenkauf, BM; Miner, B; Newton, AL; Gaydos, JK; Wynne, J; Groves, CL; Hendler, G; Murray, M; Fradkin, S; Breitbart, M; Fahsbender, E; Lafferty, KD; Kilpatrick, AM; Miner, CM; Raimondi, P; Lahner, L; Friedman, CS; Daniels, S; Haulena, M; Marliave, J; Burge, CA; Eisenlord, ME; Harvell, CD (2014). "Densovirus associated with sea-star wasting disease and mass mortality". Proc Natl Acad Sci U S A. 111 (48): 17278–83. Bibcode:2014PNAS..11117278H. doi:10.1073/pnas.1416625111. PMC 4260605. PMID 25404293.
- Cotmore SF, Agbandje-Mckenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ (2014). "The family Parvoviridae". Arch. Virol. 159 (5): 1239–47. doi:10.1007/s00705-013-1914-1. PMC 4013247. PMID 24212889.
- "2013.001a-aaaV.A.v4.Parvoviridae.pdf". Retrieved 20 February 2016.
- "ICTV 10th Report (2018)Dependoparvovirus".
- Canuti, M; Williams, CV; Gadi, SR; Jebbink, MF; Oude Munnink, BB; Jazaeri Farsani, SM; Cullen, JM; van der Hoek, L (2014). "Persistent viremia by a novel parvovirus in a slow loris (Nycticebus coucang) with diffuse histiocytic sarcoma". Front Microbiol. 5: 655. doi:10.3389/fmicb.2014.00655. PMC 4249460. PMID 25520709.
- "ICTV 10th Report (2018)".
- "ICTV 10th Report (2018)Erythroparvovirus".
- "ICTV 10th Report (2018)Protoparvovirus".
- Parker JS, Murphy WJ, Wang D, O'Brien SJ, Parrish CR (2001). "Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells". Journal of Virology. 75 (8): 3896–3902. doi:10.1128/JVI.75.8.3896-3902.2001. PMC 114880. PMID 11264378.
- Ross SR, Schofield JJ, Farr CJ, Bucan M (2002). "Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus". Proceedings of the National Academy of Sciences of the United States of America. 99 (19): 12386–90. Bibcode:2002PNAS...9912386R. doi:10.1073/pnas.192360099. PMC 129454. PMID 12218182.
- Hueffer K, Parker JS, Weichert WS, Geisel RE, Sgro JY, Parrish CR (2003). "The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor". Journal of Virology. 77 (3): 1718–26. doi:10.1128/jvi.77.3.1718-1726.2003. PMC 140992. PMID 12525605.
- Goodman LB, Lyi SM, Johnson NC, Cifuente JO, Hafenstein SL, Parrish CR (2010). "Binding site on the transferrin receptor for the parvovirus capsid and effects of altered affinity on cell uptake and infection". Journal of Virology. 84 (10): 4969–78. doi:10.1128/jvi.02623-09. PMC 2863798. PMID 20200243.
- Harbison CE, Lyi SM, Weichert WS, Parrish CR (2009). "Early steps in cell infection by parvoviruses: host-specific differences in cell receptor binding but similar endosomal trafficking". Journal of Virology. 83 (20): 10504–14. doi:10.1128/jvi.00295-09. PMC 2753109. PMID 19656887.
- Nelson CD, Minkkinen E, Bergkvist M, Hoelzer K, Fisher M, Bothner B, Parrish CR (2008). "Detecting small changes and additional peptides in the canine parvovirus capsid structure". Journal of Virology. 82 (21): 10397–407. doi:10.1128/jvi.00972-08. PMC 2573191. PMID 18701590.
- White DO, Fenner F (1994). "17 Parvoviridae: Parvovirus B19". Medical Virology (4th ed.). Academic Press. pp. 288–291. ISBN 978-0-12-746642-2.
- Fonseca, Eduardo K.U.N. (January 2018). "Etymologia: Parvovirus". Emerg Infect Dis. 24 (2): 293. doi:10.3201/eid2402.ET2402. PMC 5782889.
cites public domain text from the CDC, product of the US Government.
- Kilham L, Olivier LJ (1959). "A latent virus of rats isolated in tissue culture". Virology. 7 (4): 428–437. doi:10.1016/0042-6822(59)90071-6. PMID 13669314.
- Heegaard ED, Brown KE (2002). "Human parvovirus B19". Clin Microbiol Rev. 15 (3): 485–505. doi:10.1128/cmr.15.3.485-505.2002. PMC 118081. PMID 12097253.
Further reading
External links
| Wikimedia Commons has media related to Parvoviridae. |
| Wikispecies has information related to Parvoviridae |