Orthobunyavirus
Orthobunyavirus is a genus of the Peribunyaviridae family in the order Bunyavirales. There are currently ~170 viruses recognised in this genus. These have been assembled into 88 species[1] and 20 serogroups.
| Orthobunyavirus | |
|---|---|
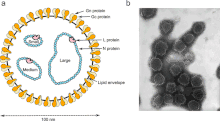 | |
| Orthobunyavirus structure (left); transmission electron micrograph of California encephalitis virus (right) | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Ellioviricetes |
| Order: | Bunyavirales |
| Family: | Peribunyaviridae |
| Genus: | Orthobunyavirus |
| Type species | |
| Bunyamwera orthobunyavirus | |
| Species[1] | |
| |
The name Orthobunyavirus derives from Bunyamwera, Uganda,[2] where the original type species Bunyamwera orthobunyavirus was first discovered,[3] along with the prefix orthos (ορϑοϛ) meaning 'straight.'[4]
Epidemiology
The genus is most diverse in Africa, Australia and Oceania, but occurs almost worldwide. Most orthobunyavirus species are transmitted by gnats and cause diseases of cattle. The California encephalitis virus, the La Crosse virus and the Jamestown Canyon virus are North American species that cause encephalitis in humans.
Virology
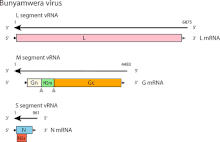
- The type species is Bunyamwera orthobunyavirus.[5]
- The virus is spherical, diameter 80 nm to 120 nm, and comprises three negative-sense single stranded RNA molecules encapsulated in a ribonucleocapsid.[6]
- The three RNAs are described as S, M and L (for Small, Medium and Large) and are circa 1kb (kilobases), 4.5kb and 6.9kb in length[7][6][8]
- The S RNA encodes the Nucleocapsid protein (N protein) and a non structural protein (NS Protein).[9]
- The M RNA encodes a polyprotein which is cleaved by host protease into Gn, NSm and Gc proteins.[6]
- The L RNA encodes the viral RNA dependent RNA Polymerase or L Protein[10]
Life cycle
Vectors
The primary vectors of Orthobunyaviruses are hematophagous insects of the Culicidae family, including members from a number of mosquito genera (including Aedes, Coquillettidia, Culex, Culiseta, and Anopheles) and biting midges (such as Culicoides paraensis).[11][8] Although transmission by ticks and bed bugs may also occur. Viral vector preference is generally strict, with only a one or very small number of vectors transmitting a specific virus in the region, even where multiple viruses and vectors overlap.[12] Organisms related to the preferential vector may be able to carry a virus but not competently transmit it.[8]
The vector arthropod acquires the virus while taking a blood meal from an infected host. In mosquitoes, replication of orthobunyaviruses is enhanced by immune modulation that occurs as a result of blood protein digestion producing GABA and the activation of GABAergic signalling.[13] Infection is transmitted to a new host via viral particles in vector saliva.[13] Orthobunyavirus infection in arthropod cells is not fully understood, but is generally non-cytopathological and deleterious effects are minimal.[14][12] Infected mosquitoes may experience an increase in fitness.[12] Transorvarial transmission has been observed among mosquitoes infected with orthobunyaviruses of the California serogroup[8] Like mosquitoes, only female culicoid midges feed on blood; they prefer indoor feeding particularly during rain.[8]
Sylvatic Cycle Hosts
In the slyvatic cycle, viruses are transmitted between mammalian hosts by the arthropod vector. A diverse range of mammals have been identified or implicated as hosts or reservoirs of orthobunyaviruses including: non-human primates, sloths, wild and domestic birds, marmosets, rodents, and large mammals such as deer, moose, and elk.[11][8]
Infection
Infection begins with the bite of an infected competent vector organism. Viral entry proceeds by receptor-mediated (clathrin-dependent) endocytosis, but which receptors unknown.[14] Although, Heparan sulfate and DC-SIGN (CD209 or Dendritic cell-specific intracellular adhesion molecule-3-grabbing non-integrin) have been identified as viral entry components in some orthobunyaviruses.[12][14] Gn/Gc heterodimers on the viral surface are responsible for target cell recognition[15], with Gc is considered the primary attachment protein, although Gn has been suggested as the attachment protein for LACV in arthropod cells.[12] Acidification of the endosome triggers a conformational change in the Gc fusion peptide, uncoating the ribonuclearprotein (RNP) as it is released into the cytoplasm.[15]
Upon release into the cytoplasm, primary transcription begins with an endonuclease domain on L protein engaging in a process known as "cap-snatching."[12][15] During cap-snatching, 10-18 nucleotides of 5' 7-methylguanylate primers are cleaved from host mRNAs and attached to prime the 5' end of the viral RNAs.[8] Like all negative-sense RNA viruses, orthobunyaviruses require ongoing, concurrent translation by the host cell to produce full-length viral mRNAs, consequently the 3' end of orthobunyavirus mRNAs lack polyadenylation.[8] Notably they are also missing the signal for polyadenylation; instead the 3' ends are thought to form a stem-loop structure.[8][12] Antigenomes (full length positive-sense RNAs) used as templates for replication of the viral genome are produced by L protein RdRp without the need for primers.[8] Both negative-sense genomes and positive-sense antigenomes are associated with N proteins (forming RNPs) at all times during the replication cycle.[16] Thus, N and L are the minimum proteins required for transcription and replication[15][12]
The M genome segment codes for the Gn-NSm-Gc polyprotein on a single open-reading frame (ORF) which is cotranslationally cleaved by internal signal peptides and host signal peptidase.[15][8] The free glycoproteins Gc and Gn insert into the membrane of the endoplasmic reticulum and form heterodimers. A Golgi retention signal on Gn, permits transport of the heterodimers to the Golgi apparatus, where glycosylation occurs. The presence of the viral glycoproteins modifies the Golgi membrane to enable budding of RNPs into a Golgi derived tubular viral factory (viroplasm).[14][8] As segmented viruses, orthobuynaviruses require precise packaging of one of each of the three genomic segments into the final virion to produce a mature, infectious particle. Packaging appears to be directed by signals contained entirely within UTR sequences.[12] The packaged genomes acquire a lipid membrane as they bud into the viral factories, are then transported to the host cell plasma membrane and released via exocytosis. A final gylcoprotein modification upon release produces a mature, infectious particle.[12]
Evolution
Orthobunyaviruses evolve partly by a key mechanism known as genomic reassortment, which also occurs in other segmented viruses. When viruses of the same group co-infect a host cell, mixtures and novel combinations of the S, M, and L segments can be produced, increasing diversity. The most common reassortment events are with the L and S segments.[17]
Serogroups
The taxonomy remains somewhat fluid as relatively few viral genomes in this genus have been sequenced. Several of the viruses listed have been shown to be recombinants of other viruses and may be reclassified.
18 serogroups have been recognized on the basis of the results of cross-hemagglutination inhibition and antibody neutralization relationships. Another - Wyeomyia - has since been recognised. Several viruses have not yet been classified into one of the serogroups.
The Simbu serogroup is the largest and contains at least 25 members. There are at least 13 members in the Group C serogroup.
Medically important viruses belong to the Bwamba, Bunyamwera, California, Group C and Simbu serogroups.
Anopheles A serogroup
- Anopheles A virus
- Tacaiuma virus
- Virgin River virus
- Trombetas complex
- Arumateua virus
- Caraipé virus
- Trombetas virus
- Tucuruí virus
Anopheles B serogroup
- Anopheles B virus
- Boraceia virus
Bakau serogroup
- Bakau virus
- Nola virus
Bunyamwera serogroup
- Birao virus
- Bozo virus
- Bunyamwera virus
- Cache Valley virus
- Fort Sherman virus
- Germiston virus
- Guaroa virus
- Ilesha virus
- Kairi virus
- Maguari virus
- Main Drain virus
- Lokern virus
- Northway virus
- Playas virus
- Potosi virus
- Shokwe virus
- Stanfield virus
- Tensaw virus
- Xingu virus
- Batai complex
- Batai virus
- Čalovo virus
- Chittoor virus
- Ngari complex
- Garissa virus
- KV-141 virus
- Ngari virus
Bwamba serogroup
- Bwamba virus
- Pongola virus
California serogroup
California encephalitis virus
Chatanga virus
Inkoo virus
Jamestown Canyon virus
Jerry Slough virus
Keystone virus
Khatanga virus
La Crosse virus
Lumbo virus
Melao virus
Morro Bay virus
San Angelo virus
Serra do Navio virus
Snowshoe hare virus
South River virus
Tahyna virus
Trivittatus virus
Capim serogroup
Acara virus
Benevides virus
Capim virus
Gamboa serogroup
- Alajuela virus
- Gamboa virus
- 75V 2621 virus strain
- Pueblo Viejo virus
- San Juan virus
Group C serogroup
Bruconha virus
Ossa virus
Caraparu complex
- Apeu virus
- Bruconha virus
- Caraparu virus
- Vinces virus
Madrid complex
- Madrid virus
- Marituba complex
- Gumbo limbo virus
- Marituba virus
- 63U-11 virus strain
- Murutucu virus
- Nepuyo virus
- Restan virus
Oriboca complex
- Itaqui virus
- Oriboca virus
Guama serogroup
Ananindeua virus
Bertioga virus
Bimiti virus
Cananeia virus
Catu virus
Gan Gan virus
Guama virus
Guaratuba virus
Itimirim virus
Mahogany hammock virus
Mirim virus
Timboteua virus
Trubanaman virus
Koongol serogroup
Koongol virus
Wongal virus
Mapputta serogroup
Buffalo Creek virus
Mapputta virus
Maprik virus
Murrumbidgee virus
Salt Ash virus
Minatitlan serogroup
Minatitlan virus
Palestina virus
Nyando serogroup
Eretmapodites virus
Nyamdo virus
Olifanstlei serogroup
Botambi virus
Olifanstlei virus
Patois serogroup
Abras virus
Babahoyo virus
Pahayokee virus
Patois virus
Shark River virus
Zegla virus
Simbu serogroup
Iquitos virus
Jatobal virus
Leanyer virus
Mermet virus
Oya virus
Thimiri virus
Akabane serocomplex
- Akabane virus
- Tinaroo virus
Oropouche serocomplex
- Madre de Dios virus (MDDV)
- Oropouche virus
Sathuperi serocomplex
- Douglas virus
- Sathuperi virus
Shamonda serocomplex
- Peaton virus
- Shamonda virus
Shuni serocomplex
- Aino virus
- Shuni virus
Simbu complex
- Schmallenberg virus[18]
- Simbu virus
Tete serogroup
Bahig virus
Batama virus
Matruh virus
Tete virus
Tsuruse virus
Weldona virus
Turlock serogroup
Kedah virus
Lednice virus
M'Poko virus
Turlock virus
Umbre virus
Wyeomyia serogroup
Anhembi virus
Cachoeira Porteira virus
Iaco virus
Macaua virus
Sororoca virus
Taiassui virus
Tucunduba virus
Wyeomyia virus
Unclassified
Batama virus
Bellavista virus
Belmont virus
Enseada virus
Estero Real virus
Herbert virus
Jonchet virus
Jurona virus
Kaeng Khei virus
Kibale virus
Kowanyama virus
Mojuí dos Campos virus
Ntwetwe virus[19]
Taï virus
Tataguine virus
Triniti virus
Witwatersrand virus
Wolkberg virus
Yacaaba virus
References
- "Virus Taxonomy: 2019 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 6 May 2020.
- "ICTV 9th Report (2011) Bunyaviridae" (html). International Committee on Taxonomy of Viruses (ICTV). Retrieved 31 January 2019.
Bunya: from Bunyamwera, place in Uganda, where type virus was isolated.
- Smithburn KC, Haddow AJ, Mahaffy AF (March 1946). "A neurotropic virus isolated from Aedes mosquitoes caught in the Semliki forest". The American Journal of Tropical Medicine and Hygiene. 26 (2): 189–208. doi:10.4269/ajtmh.1946.s1-26.189. OCLC 677158400. PMID 21020339.
- Griffith C (2005). "Dictionary of Botanical Epithets". Dictionary of Botanical Epithets. Retrieved 31 January 2019.
orthos orth adj ορϑοϛ straight
- "International Committee on Taxonomy of Viruses, Bunyaviridae". bio-mirror.cn. Archived from the original on 9 September 2013. Retrieved 20 December 2018.
- "ViralZone page". viralzone.expasy.org. Retrieved 20 December 2018.
- Kascsak RJ, Lyons MJ (October 1977). "Bunyamwera virus. I. The molecular complexity of the virion RNA". Virology. 82 (1): 37–47. doi:10.1016/0042-6822(77)90030-7. PMID 898678.
- Evans AB, Peterson KE (August 2019). "Throw out the Map: Neuropathogenesis of the Globally Expanding California Serogroup of Orthobunyaviruses". Viruses. 11 (9): 794. doi:10.3390/v11090794. PMC 6784171. PMID 31470541.
- Genbank: Bunyamwera virus segment S, complete sequence
- Genbank: Bunyamwera virus L segment, complete sequence
- Sakkas H, Bozidis P, Franks A, Papadopoulou C (April 2018). "Oropouche Fever: A Review". Viruses. 10 (4): 175. doi:10.3390/v10040175. PMC 5923469. PMID 29617280.
- Elliott RM (October 2014). "Orthobunyaviruses: recent genetic and structural insights". Nature Reviews. Microbiology. 12 (10): 673–85. doi:10.1038/nrmicro3332. PMID 25198140.
- Wu P, Yu X, Wang P, Cheng G (March 2019). "Arbovirus lifecycle in mosquito: acquisition, propagation and transmission". Expert Reviews in Molecular Medicine. 21: e1. doi:10.1017/erm.2018.6. PMID 30862324.
- Dutuze MF, Nzayirambaho M, Mores CN, Christofferson RC (2018-04-12). "Orthobunyaviruses With Potential One Health Implications". Frontiers in Veterinary Science. 5: 69. doi:10.3389/fvets.2018.00069. PMC 5906542. PMID 29707545.
- Pawaiya RV, Gupta VK (2013-11-21). "A review on Schmallenberg virus infection: a newly emerging disease of cattle, sheep and goats". Veterinární Medicína. 58 (10): 516–526. doi:10.17221/7083-vetmed. ISSN 0375-8427.
- Zheng W, Tao YJ (May 2013). "Genome encapsidation by orthobunyavirus nucleoproteins". Proceedings of the National Academy of Sciences of the United States of America. 110 (22): 8769–70. Bibcode:2013PNAS..110.8769Z. doi:10.1073/pnas.1306838110. PMC 3670359. PMID 23696659.
- da Rosa, Jorge Fernando Travassos; de Souza, William Marciel; de Paula Pinheiro, Francisco; Figueiredo, Mário Luiz; Cardoso, Jedson Ferreira; Acrani, Gustavo Olszanski; Nunes, Márcio Roberto Teixeira (2017-02-06). "Oropouche Virus: Clinical, Epidemiological, and Molecular Aspects of a Neglected Orthobunyavirus". The American Journal of Tropical Medicine and Hygiene. 96 (5): 16–0672. doi:10.4269/ajtmh.16-0672. ISSN 0002-9637. PMC 5417190. PMID 28167595.
- Nachrichten, n-tv. "Unbekanntes Virus entdeckt". n-tv.de. Retrieved 20 December 2018.
- Edridge AW, Deijs M, Namazzi R, Cristella C, Jebbink MF, Maurer I, et al. (January 2019). "Novel Orthobunyavirus Identified in the Cerebrospinal Fluid of a Ugandan Child With Severe Encephalopathy". Clinical Infectious Diseases. 68 (1): 139–142. doi:10.1093/cid/ciy486. PMC 6293039. PMID 29893821.