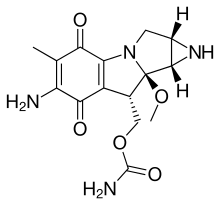
Mitomycins
The mitomycins are a family of aziridine-containing natural products isolated from Streptomyces caespitosus or Streptomyces lavendulae.[1][2] They include mitomycin A, mitomycin B, and mitomycin C. When the name mitomycin occurs alone, it usually refers to mitomycin C, its international nonproprietary name. Mitomycin C is used as a medicine for treating various disorders associated with the growth and spread of cells.
Biosynthesis
In general, the biosynthesis of all mitomycins proceeds via combination of 3-amino-5-hydroxybenzoic acid (AHBA), D-glucosamine, and carbamoyl phosphate, to form the mitosane core, followed by specific tailoring steps.[3] The key intermediate, AHBA, is a common precursor to other anticancer drugs, such as rifamycin and ansamycin.
Specifically, the biosynthesis begins with the addition of phosphoenolpyruvate (PEP) to erythrose-4-phosphate (E4P) with a yet undiscovered enzyme, which is then ammoniated to give 4-amino-3-deoxy-D-arabino heptulosonic acid-7-phosphate (aminoDHAP). Next, DHQ synthase catalyzes a ring closure to give 4-amino3-dehydroquinate (aminoDHQ), which then undergoes a double oxidation via aminoDHQ dehydratase to give 4-amino-dehydroshikimate (aminoDHS). The key intermediate, 3-amino-5-hydroxybenzoic acid (AHBA), is made via aromatization by AHBA synthase.
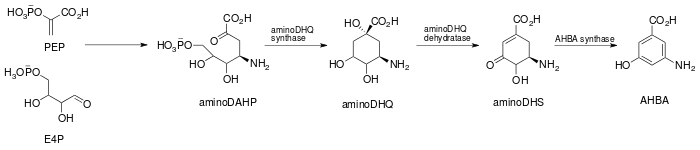
Synthesis of the key intermediate, 3-amino-5-hydroxy-benzoic acid.
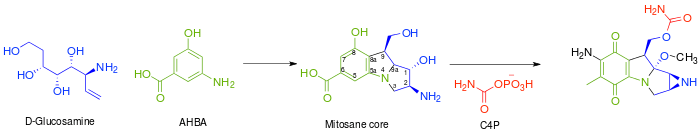
The mitosane core is synthesized as shown below via condensation of AHBA and D-glucosamine, although no specific enzyme has been characterized that mediates this transformation. Once this condensation has occurred, the mitosane core is tailored by a variety of enzymes. Both the sequence and the identity of these steps are yet to be determined.
- Complete reduction of C-6 – Likely via F420-dependent tetrahydromethanopterin (H4MPT) reductase and H4MPT:CoM methyltransferase
- Hydroxylation of C-5, C-7 (followed by transamination), and C-9a. – Likely via cytochrome P450 monooxygenase or benzoate hydroxylase
- O-Methylation at C-9a – Likely via SAM dependent methyltransferase
- Oxidation at C-5 and C8 – Unknown
- Intramolecular amination to form aziridine – Unknown
- Carbamoylation at C-10 – Carbamoyl transferase, with carbamoyl phosphate (C4P) being derived from L-citrulline or L-arginine
Biological effects
In the bacterium Legionella pneumophila, mitomycin C induces competence for transformation.[4] Natural transformation is a process of DNA transfer between cells, and is regarded as a form of bacterial sexual interaction. In the fruit fly Drosophila melanogaster, exposure to mitomycin C increases recombination during meiosis, a key stage of the sexual cycle.[5] In the plant Arabidopsis thaliana, mutant strains defective in genes necessary for recombination during meiosis and mitosis are hypersensitive to killing by mitomycin C.[6]
Medicinal uses and research
Mitomycin C has been shown to have activity against stationary phase persisters caused by Borrelia burgdorferi, a factor in lyme disease.[7][8] Mitomycin C is used to treat symptoms of pancreatic and stomach cancer,[9] and is under clinical research for its potential to treat gastrointestinal strictures,[10] wound healing from glaucoma surgery,[11] corneal excimer laser surgery[12] and endoscopic dacryocystorhinostomy.[13]
References
- Clokie, Martha R. J.; Kropinski, Andrew M. (Andrew Maitland Boleslaw) (2009). Bacteriophages : methods and protocols. Humana Press. ISBN 9781603271646. OCLC 297169927.
- Danshiitsoodol N, de Pinho CA, Matoba Y, Kumagai T, Sugiyama M (2006). "The mitomycin C (MMC)-binding protein from MMC-producing microorganisms protects from the lethal effect of bleomycin: crystallographic analysis to elucidate the binding mode of the antibiotic to the protein". J Mol Biol. 360 (2): 398–408. doi:10.1016/j.jmb.2006.05.017. PMID 16756991.
- Mao Y.; Varoglu M.; Sherman D.H. (April 1999). "Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces Iavendulae NRRL 2564". Chemistry and Biology. 6 (4): 251–263. doi:10.1016/S1074-5521(99)80040-4. PMID 10099135.
- Charpentier X, Kay E, Schneider D, Shuman HA (March 2011). "Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila". J. Bacteriol. 193 (5): 1114–21. doi:10.1128/JB.01146-10. PMC 3067580. PMID 21169481.
- Schewe MJ, Suzuki DT, Erasmus U (July 1971). "The genetic effects of mitomycin C in Drosophila melanogaster. II. Induced meiotic recombination". Mutat. Res. 12 (3): 269–79. doi:10.1016/0027-5107(71)90015-7. PMID 5563942.
- Bleuyard JY, Gallego ME, Savigny F, White CI (February 2005). "Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair". Plant J. 41 (4): 533–45. doi:10.1111/j.1365-313X.2004.02318.x. PMID 15686518.
- Feng, Jie; Shi, Wanliang; Zhang, Shuo; Zhang, Ying (3 June 2015). "Identification of new compounds with high activity against stationary phase Borrelia burgdorferi from the NCI compound collection". Emerging Microbes & Infections. 4 (5): e31. doi:10.1038/emi.2015.31. PMC 5176177. PMID 26954881.
- Sharma, Bijaya; Brown, Autumn V.; Matluck, Nicole E.; Hu, Linden T.; Lewis, Kim (26 May 2015). "Borrelia burgdorferi, the Causative Agent of Lyme Disease, Forms Drug-Tolerant Persister Cells". Antimicrobial Agents and Chemotherapy. 59 (8): AAC.00864–15. doi:10.1128/AAC.00864-15. PMC 4505243. PMID 26014929.
- "Mitomycin". Drugs.com. 2017. Retrieved 11 November 2017.
- Rustagi, T; Aslanian, H. R; Laine, L (2015). "Treatment of Refractory Gastrointestinal Strictures with Mitomycin C: A Systematic Review". Journal of Clinical Gastroenterology. 49 (10): 837–47. doi:10.1097/MCG.0000000000000295. PMID 25626632.
- Cabourne, E; Clarke, J. C; Schlottmann, P. G; Evans, J. R (2015). "Mitomycin C versus 5-fluorouracil for wound healing in glaucoma surgery" (PDF). The Cochrane Database of Systematic Reviews (11): CD006259. doi:10.1002/14651858.CD006259.pub2. PMID 26545176.
- Majmudar, Parag A; Forstot, S.Lance; Dennis, Richard F; Nirankari, Verinder S; Damiano, Richard E; Brenart, Robert; Epstein, Randy J (January 2000). "Topical Mitomycin-C for subepithelial fibrosis after refractive corneal surgery". Ophthalmology. 107 (1): 89–94. doi:10.1016/s0161-6420(99)00019-6. ISSN 0161-6420. PMID 10647725.
- Cheng, S. M; Feng, Y. F; Xu, L; Li, Y; Huang, J. H (2013). "Efficacy of Mitomycin C in Endoscopic Dacryocystorhinostomy: A Systematic Review and Meta-Analysis". PLoS ONE. 8 (5): e62737. Bibcode:2013PLoSO...862737C. doi:10.1371/journal.pone.0062737. PMC 3652813. PMID 23675423.