Molekel
Molekel is a free software multiplatform molecular visualization program.[1] It was originally developed at the University of Geneva by Peter F. Flükiger in the 1990s for Silicon Graphics Computers. In 1998, Stefan Portmann took over responsibility and released Version 3.0. Version 4.0 was a nearly platform independent version. Further developments lead to version 4.3, before Stefan Portmann moved on and ceased to develop the codes. In 2006, the Swiss National Supercomputing Centre (CSCS) restarted the project and version 5.0 was released on 21 December 2006.[2]
| Developer(s) | Swiss National Supercomputing Centre |
|---|---|
| Stable release | 5.4
/ August 2009 |
| Operating system | Linux, Microsoft Windows, Mac OS X |
| Type | Molecular modelling |
| License | GNU General Public License |
| Website | ugovaretto |
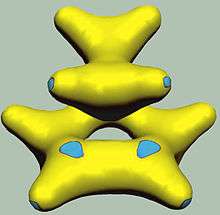
Electron density plot of the norbornyl cation determined using DFT with a mapping of mulliken electrostatic potential- B3LYP/6-31G* in Gaussian03, picture drawn in molekel using Gaussian cube file; Positive charge is colored blue in this picture.
Major features
- Visualization of residues (ribbon or schematic)
- Complete control over the generation of molecular surfaces (bounding box and resolution)
- Visualization of the following surfaces:
- orbitals
- Isosurface from electron density data
- Isosurface from Gaussian cube grid data
- Solvent-accessible surface (SAS)
- Solvent excluded surface (SES)
- Van der Waals radii
- Animation of molecular surfaces
- Export to PostScript or TIFF
gollark: It's the same file.
gollark: How is anyone getting these URLs?
gollark: `htech-obliterating-memetic-hazards.tar.zst` sounds fun.
gollark: Also an old copy of FractalArt.
gollark: Oh, apparently this contains a 1.9GB SQL dump from the webcrawler too?
See also
- Gabedit
- List of molecular graphics systems
- Molden
- Molecular graphics
- Software for molecular mechanics modeling
- SAMSON
- List of free and open-source software packages
References
- Noel O'Blog
- Molekel About Archived 2009-08-27 at the Wayback Machine
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.