Accessible surface area
The accessible surface area (ASA) or solvent-accessible surface area (SASA) is the surface area of a biomolecule that is accessible to a solvent. Measurement of ASA is usually described in units of square Ångstroms (a standard unit of measurement in molecular biology). ASA was first described by Lee & Richards in 1971 and is sometimes called the Lee-Richards molecular surface.[1] ASA is typically calculated using the 'rolling ball' algorithm developed by Shrake & Rupley in 1973.[2] This algorithm uses a sphere (of solvent) of a particular radius to 'probe' the surface of the molecule.
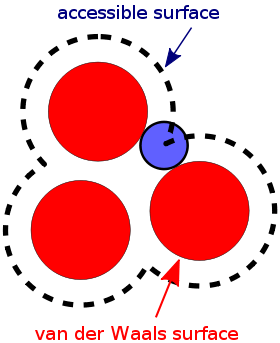
Methods of calculating ASA
Shrake-Rupley algorithm
The Shrake-Rupley algorithm is a numerical method that draws a mesh of points equidistant from each atom of the molecule and uses the number of these points that are solvent accessible to determine the surface area.[2] The points are drawn at a water molecule's estimated radius beyond the van der Waals radius, which is effectively similar to ‘rolling a ball’ along the surface. All points are checked against the surface of neighboring atoms to determine whether they are buried or accessible. The number of points accessible is multiplied by the portion of surface area each point represents to calculate the ASA. The choice of the 'probe radius' does have an effect on the observed surface area, as using a smaller probe radius detects more surface details and therefore reports a larger surface. A typical value is 1.4Å, which approximates the radius of a water molecule. Another factor that affects the results is the definition of the VDW radii of the atoms in the molecule under study. For example, the molecule may often lack hydrogen atoms which are implicit in the structure. The hydrogen atoms may be implicitly included in the atomic radii of the 'heavy' atoms, with a measure called the 'group radii'. In addition, the number of points created on the van der Waals surface of each atom determines another aspect of discretization, where more points provide an increased level of detail.
LCPO method
The LCPO method uses a linear approximation of the two-body problem for a quicker analytical calculation of ASA.[3] The approximations used in LCPO result in an error in the range of 1-3 Ų.
Power Diagram method
Recently a method was presented that calculates ASA fast and analytically using a power diagram.[4]
Applications
Accessible surface area is often used when calculating the transfer free energy required to move a biomolecule from aqueous solvent to a non-polar solvent such as a lipid environment. The LCPO method is also used when calculating implicit solvent effects in the molecular dynamics software package AMBER.
It is recently suggested that (predicted) accessible surface area can be used to improve prediction of protein secondary structure.[5][6]
Relation to solvent-excluded surface
The ASA is closely related to the concept of the solvent-excluded surface (also known as the molecular surface or Connolly surface), which is imagined as a cavity in bulk solvent (effectively the inverse of the solvent-accessible surface). It is also calculated in practice via a rolling-ball algorithm developed by Frederic Richards[7] and implemented three-dimensionally by Michael Connolly in 1983[8] and Tim Richmond in 1984.[9] Connolly spent several more years perfecting the method.[10]
See also
- Implicit solvation
- Van der Waals surface
- VADAR tool for analyzing peptide and protein structures
- Relative accessible surface area
Notes
- Lee, B; Richards, FM. (1971). "The interpretation of protein structures: estimation of static accessibility". J Mol Biol. 55 (3): 379–400. doi:10.1016/0022-2836(71)90324-X. PMID 5551392.
- Shrake, A; Rupley, JA. (1973). "Environment and exposure to solvent of protein atoms. Lysozyme and insulin". J Mol Biol. 79 (2): 351–71. doi:10.1016/0022-2836(73)90011-9. PMID 4760134.
- Weiser J, Shenkin PS, Still WC (1999). "Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO)". Journal of Computational Chemistry. 20 (2): 217–230. doi:10.1002/(SICI)1096-987X(19990130)20:2<217::AID-JCC4>3.0.CO;2-A.
- Klenin K, Tristram F, Strunk T, Wenzel W (2011). "Derivatives of molecular surface area and volume: Simple and exact analytical formulas". Journal of Computational Chemistry. 32 (12): 2647–2653. doi:10.1002/jcc.21844. PMID 21656788.
- Momen-Roknabadi, A; Sadeghi, M; Pezeshk, H; Marashi, SA (2008). "Impact of residue accessible surface area on the prediction of protein secondary structures". BMC Bioinformatics. 9: 357. doi:10.1186/1471-2105-9-357. PMC 2553345. PMID 18759992.
- Adamczak, R; Porollo, A; Meller, J. (2005). "Combining prediction of secondary structure and solvent accessibility in proteins". Proteins. 59 (3): 467–75. doi:10.1002/prot.20441. PMID 15768403.
- Richards, FM. (1977). "Areas, volumes, packing and protein structure". Annu Rev Biophys Bioeng. 6: 151–176. doi:10.1146/annurev.bb.06.060177.001055. PMID 326146.
- Connolly, M. L. (1983). "Analytical molecular surface calculation". J Appl Crystallogr. 16 (5): 548–558. doi:10.1107/S0021889883010985.
- Richmond, T. J. (1984). "Solvent accessible surface area and excluded volume in proteins. Analytical equations for overlapping spheres and implications for the hydrophobic effect". J Mol Biol. 178 (1): 63–89. doi:10.1016/0022-2836(84)90231-6. PMID 6548264.
- Connolly, M. L. (1993). "The molecular surface package". J Mol Graphics. 11 (2): 139–141. doi:10.1016/0263-7855(93)87010-3.
References
- Connolly, M. L. (1983). "Solvent-accessible surfaces of proteins and nucleic-acids". Science. 221 (4612): 709–713. Bibcode:1983Sci...221..709C. doi:10.1126/science.6879170.
- Richmond, Timothy J. (1984). "solvent accessible surface area and excluded volume in proteins". J. Mol. Biol. 178: 63–89. doi:10.1016/0022-2836(84)90231-6. PMID 6548264.
- Connolly, Michael L. (1985). "Computation of molecular volume". J. Am. Chem. Soc. 107 (5): 118–1124. doi:10.1021/ja00291a006.
- Connolly, M. L. (1991). "Molecular interstitial skeleton". Computers and Chemistry. 15 (1): 37–45. doi:10.1016/0097-8485(91)80022-E.
- Sanner, M.F. (1992). Modelling and Applications of Molecular Surfaces (PhD thesis).
- Connolly, M. L. (1992). "Shape distributions of protein topography". Biopolymers. 32 (9): 1215–1236. doi:10.1002/bip.360320911. PMID 1420989.
- Blaney, J. M. (1994). "Distance Geometry in Molecular Modeling". Reviews in Computational Chemistry. Rev. Comput. Chem. pp. 299–335. doi:10.1002/9780470125823.ch6. ISBN 9780470125823.
- Grant, J. A.; Pickup, B. T. (1995). "A Gaussian description of molecular shape". J. Phys. Chem. 99 (11): 3503–3510. doi:10.1021/j100011a016.
- Boissonnat, Jean-Daniel; Devillers, Olivier; Duquesne, Jacqueline; Yvinec, Mariette (1994). "Computing Connolly Surfaces". Journal of Molecular Graphics. 12 (1): 61–62. doi:10.1016/0263-7855(94)80033-2. ISSN 1093-3263.
- Petitjean, M (1994). "On the Analytical Calculation of van der Waals Surfaces and Volumes: Some Numerical Aspects". J. Comput. Chem. 15 (5). pp. 507–523. doi:10.1002/jcc.540150504.
- Connolly, M. L. (1996). "Molecular Surfaces: A Review". Network Science. Archived from the original on 2013-03-15. Cite journal requires
|journal=(help) - Lin, S. L. (1994). "Molecular surface representations by sparse critical points". Proteins. 18 (1): 94–101. doi:10.1002/prot.340180111. PMID 8146125.
- Gerstein, M; Richards, F.S. (2001). "Protein geometry: Volumes, areas and distances". CiteSeerX 10.1.1.134.2539. Cite journal requires
|journal=(help) - Voss, N. R. (2006). "The geometry of the ribosomal polypeptide exit tunnel". J. Mol. Biol. 360 (4): 893–906. CiteSeerX 10.1.1.144.6548. doi:10.1016/j.jmb.2006.05.023. PMID 16784753.
- Leach, A. (2001). Molecular Modelling: Principles and Applications (2nd ed.). p. 7.
- Busa, Jan; Dzurina, Jozef; Hayryan, Edik (2005). "ARVO: A fortran package for computing the solvent accessible surface area and the excluded volume of overlapping spheres via analytic equations". Comput. Phys. Commun. 165 (1): 59–96. Bibcode:2005CoPhC.165...59B. doi:10.1016/j.cpc.2004.08.002.
External links
- Network Science, Part 5: Solvent-Accessible Surfaces
- AREAIMOL is a command line tool in the CCP4 Program Suite for calculating ASA.
- NACCESS solvent accessible area calculations.
- FreeSASA Open source command line tool, C library and Python module for calculating ASA.
- Surface Racer Oleg Tsodikov's Surface Racer program. Solvent accessible and molecular surface area and average curvature calculation. Free for academic use.
- ASA.py — a Python-based implementation of the Shrake-Rupley algorithm.
- Michel Sanner's Molecular Surface – the fastest program to calculate the excluded surface.
- pov4grasp render molecular surfaces.
- Molecular Surface Package — Michael Connolly's program.
- Volume Voxelator — A web-based tool to generate excluded surfaces.
- ASV freeware Analytical calculation of the volume and surface of the union of n spheres (Monte-Carlo calculation also provided).
- Vorlume Computing Surface Area and Volume of a Family of 3D Balls.