MicrobesOnline
MicrobesOnline is a publicly and freely accessible website that hosts multiple comparative genomic tools for comparing microbial species at the genomic, transcriptomic and functional levels.[1][2] MicrobesOnline was developed by the Virtual Institute for Microbial Stress and Survival, which is based at the Lawrence Berkeley National Laboratory in Berkeley, California. The site was launched in 2005, with regular updates until 2011.
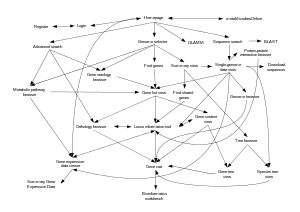
The main aim of MicrobesOnline is to provide an easy-to-use resource that integrates a wealth of data from multiple sources. This integrated platform facilitates studies in comparative genomics, metabolic pathway analysis, genome composition, functional genomics as well as in protein domain and family data. It also provides tools to search or browse the database with genes, species, sequences, orthologous groups, gene ontology (GO) terms or pathway keywords, etc. Another one of its main features is the Gene Cart, which allows users to keep a record of their genes of interest. One of the highlights of the database is the overall navigation accessibility and interconnection between the tools.
Background
The development of high-throughput methods for genome sequencing has brought about a wealth of data that requires sophisticated bioinformatics tools for their analysis and interpretation.[3] Nowadays, numerous tools exist to study genomics sequence data and extract information from different perspectives. However, the lack of unification of nomenclature and standardised protocols between tools, makes direct comparison between their results very difficult.[4] Additionally, the user is forced to constantly switch from various websites or software, adjusting the format of their data to fit with individual requirements. MicrobesOnline was developed with the aim to integrate the capacities of different tools into a unified platform for easy comparison between analysis results, with a focus on prokaryote species and basal eukaryotes.
Species included in the database
MicrobesOnline hosts genomic, gene expression and fitness data for a wide range of microbial species. Genomic data is available for 1752 bacteria, 94 archaea and 119 eukaryotes, for a total of 3707 genomes, 2842 of which are marked as being complete. Gene expression data is available for 113 species, and fitness data is available for 4 organisms.[5]
Functions and Site Architecture
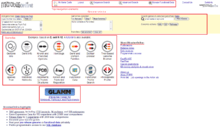
MicrobesOnline provides diverse tools for searching, analysing and integrating information related to bacteria genomes for applications in four major areas: genetic information, functional genomics, comparative genomics and metabolic pathway studies.[6] The homepage of MicrobesOnline is the portal for accessing its functions, which includes six main sections: the top navigation elements, a genome selector, examples of the tutorial based on E.coli K-12, a link to the Genome-Linked Application for Metabolic Maps (GLAMM), website highlights and the “about MicrobesOnline” list. As an ongoing project, the authors of MicrobesOnline claim that the tools for data analysis and the support of more data types will be expanded.[7]
Genetic information
Information of microbial genes stored in MicrobesOnline includes sequences (genes, transcripts and proteins), genomic loci, gene annotations and some statistics of sequences. This information can be accessed through three features displayed on the homepage of MicrobesOnline: sequence search and advanced search in the top navigation section, and the genome selector. For the sequence search tool, MicrobesOnline integrates BLAT, FastHMM and FastBLAST [8] to search protein sequences, and uses MEGABLAST to search nucleotide sequences.[9] It also provides a link to BLAST as an alternative way for searching sequences. On the other hand, the advanced search tool enables a user to search genetic information by categories, custom query, wild-card search and field-specific search, which uses the gene name, the description, the cluster of orthologous groups (COGs) id, the GO term, the KEGG enzyme commission (EC) number, etc. as key words.

The “genomes selected” box of the genome selector lists genomes added from the favourite genome list on the left or the ones searched by keywords. On the right side of the genome selector, four actions can be applied after selecting genomes: the “find genes” interface searches the gene name in the selected genomes and displays results in the gene list view; the “info” button lists a brief summary of selected genomes in the Summary View; the “GO” button opens a GO Browser called VertiGo which tabulates the number of genes under different GO items; finally, the “pathway” button initiates a pathway browser that illustrates the complete pathways of all organisms in the MicrobesOnline database.
In addition, the genome names shown in the summary view leads to a single-genome data view that presents a wealth of information about the selected genome. In the gene list view, the links “G O D H S T B...” lead the user to a locus information tool, where detailed information such as operon & regulon, domains & families, sequences, annotations, etc. are shown.
Gene carts

An important feature to store a user's work is the Gene Cart. Many web pages of MicrobesOnline displaying genetic information contain a link to add genes of interest to the session gene cart, which is available for all users. This is a temporary gene cart, and as such it loses information as a user closes the web browser. Genes in the session gene cart can be saved to the permanent gene cart which is only available to registered users after logging in.
Functional genomics
One goal of setting up MicrobesOnline is to store functional information of microbial genomes. Such information includes gene ontology and microarray-based gene expression profiles, which can be accessed through two interfaces called GO browser and Expression Data Viewer respectively. The GO browser provides links to genes organised by gene ontology terms and the Expression Data Viewer provides both the access to expression profiles and information of experimental conditions.
Gene ontology hierarchy

The GO Browser, also known as VertiGo, is used by MicrobesOnline to search and visualise the GO hierarchy, which is a unified verbal system that describes properties of gene products, including cellular components, molecular function and biological process. The Genome Selector of the MicrobesOnline homepage provides a direct way to browse the GO hierarchy of the selected genomes, as well as provide a list of genes under a selected GO term, which can then be added to the session gene cart for further analysis.
Gene expression information
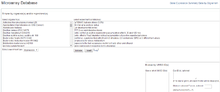
The Expression Data Viewer is an interface for searching and inspecting microarray-base gene expression experiments and expression profiles. It consists of several components: an experiment browser for searching specific experiments in selected genomes under selected experimental conditions, an expression experiment viewer providing details of each microarray experiment, a gene expression viewer showing a heat map of the expression levels of the selected gene and genes in the same operon, and finally, a profile search tool for searching gene expression profiles. The Expression Data Viewer can be accessed through three ways: the “Browse Functional Data” in the navigator bar, the “Gene Expression Data” in the homepage and the “Gene expression” list in the single-genome data view, where the expression data are available. The single-genome data view can also show a protein-protein interaction browser that allows the inspection of interaction complexes and the download of expression data (e.g. Escherichia coli str. K-12 substr. MG1655). Furthermore, the user can launch a MultiExperiment Viewer (MeV) in the single-genome data view for analysing and visualising expression data.
Comparative genomics
MicrobesOnline stores information of gene homology and phylogeny for comparative genomic studies, which can be accessed through two interfaces. The first one is the Tree Browser, which draws a species tree or a gene tree for the selected gene and its gene neighbourhood. The second one is the Orthology Browser, which is an extension of the Genome Browser and demonstrates the selected gene within the context of its gene neighbourhood aligned with orthologs in other selected genomes.[10] Both browsers provide options to save a gene in the session gene cart for further analysis.
Tree browser

The tree browser can be accessed by searching a gene by the Find Genes tool on the homepage with its VIMSS id (e.g. VIMSS15779). Once the gene context view has been accessed through the “Browse genomes by trees” option, a gene tree and a gene context diagram are displayed. In addition, the “View species tree” option opens a species tree view, which shows a species tree alongside the gene tree. Additionally, the tree browser enables users to choose both genes and genomes according to their similarity. Furthermore, it also demonstrates horizontal gene transfers among genomes.
Orthology browser

The Orthology Browser displays orthologs of genomes compared to the query genome by choosing multiple genomes from the “Select Organism(s) to Display” box.
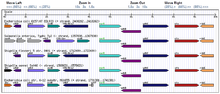
The locus information can be viewed through the “view genes” option, and this gene can be added to the session gene cart, or its gene expression data (including the heatmap) can be downloaded. Alternatively, a gene context view appears when browsing genomes by trees.
Metabolic pathway information
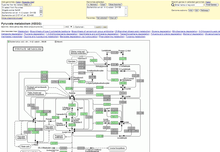
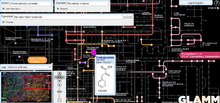
The Pathway Browser lets users to navigate the Kyoto Encyclopedia of Genes and Genomes (KEGG)[11] pathway maps displaying predicted presence or absence of enzymes for up to two selected genomes. The map of a particular pathway and a comparison between two kinds of microbes can be shown in the pathway browser. The enzyme commission number (e.g. 3.1.3.25) provides a link to the gene list view that shows information of the selected enzyme and allows the user to add genes to the session gene cart.
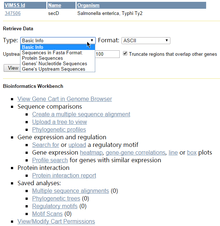
The GLAMM is another tool for searching and visualising metabolic pathways in a unified web interface. It helps users to identify or construct novel, transgenic pathways.[12]
Bioinformatics
MicrobesOnline has integrated numerous tools for analysing sequences, gene expression profiles and protein-protein interactions into an interface called Bioinformatics Workbench, which is accessed via gene carts. Analyses currently supported include multiple sequence alignments, construction of phylogenetic trees, motif searches and scans, summaries of gene expression profiles and protein-protein interactions. In order to save computational resources, a user is allowed to run two concurrent jobs for at most four hours and all results are saved temporarily until the session is terminated.[13] Results can be shared with other users or groups via the resource access control tool.
Supporting databases
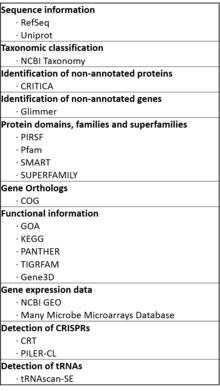
MicrobesOnline is built on the integration of the data of an array of databases that manage different aspects of its capabilities. A comprehensive list is as follows:[14]
- Sequence information: Non-redundant protein, gene and transcript sequences and annotations are extracted from RefSeq [15] and Uniprot.[16]
- Taxonomic classification of species and sequences: NCBI Taxonomy [17] is used to classify the species and sequences into phylogenetic groups, and build a phylogenetic tree.
- Identification of non-annotated proteins from sequences: CRITICA [18] is used to finds stretches of DNA sequence that code for proteins. Both a comparative genomics and a comparison and annotation-independent method are used.
- Identification of non-annotated genes from sequences: MicrobesOnline relies on Glimmer [19] to automatically find genes in bacteria, archaea and viral sequences.
- Classification of proteins: The classification of proteins by their conserved domain, family and superfamilies determined by PIRSF,[20] Pfam,[21] SMART [22] and SUPERFAMILY [23] repositories are included.
- Gene Orthology information: The orthologous groups of genes across species are based on the information on the COG database,[24] which relies on protein sequence comparison for the detection of homology.
- Functional information of genes and proteins: The range of functional information provided is contributed by the following: GOA [25] for Gene Ontology annotation of genes into functional categories, KEGG [26] for metabolic, molecular and signaling pathways of genes, and PANTHER [27][28] for information about molecular and functional pathways, in the context of the relationships between protein families and their evolution . TIGRFAMs [29] and Gene3D [30] are referred to for structural information and annotation of proteins.
- Gene expression data: Both NCBI GEO [31] and Many Microbe Microarrays Database [32] support the gene expression data of MicrobesOnline. The datasets compiled by Many Microbe Microarrays Database have the added advantage of being directly comparable, since only data generated by single-channel Affymetrix microarrays are accepted, and are subsequently normalised.
- Detection of CRISPRs: CRISPR [33] are DNA loci involved in the immunity against invasive sequences, where short direct repeats are separated by spacer sequences.[34] The databases generated by the CRT [35] and PILER-CL [36] algorithms are used to detect CRISPRs.
- Detection of tRNAs: The tRNAscan-SE [37] database is used as a reference to identify tRNA sequences.
- Submission of data by users: Users have the capacity of uploading both genomes and expression files to MicrobesOnline and analyse them with the analysis tools offered, with the option of keeping the data private (in the case of unpublished data) or releasing it to the public.[38] Microarray data should include a clear identification of the organisms, platforms, treatments and controls, experimental conditions, time points and normalization techniques used, as well as the expression data in either log ratio or log levels format. Although draft genome sequences are accepted, they must be compliant with certain guidelines: (1) the assembled genome must have less than 100 scaffolds, (2) the FASTA file format should be used, having a unique label per contig, (3) preferably gene predictions should be present (in this case, accepted formats include GenBank, EMBL, tab-delimited and FASTA), (4) the name of the genome and the NCBI taxonomy ID should be provided.
Updates
MicrobesOnline was updated every 3 to 9 months from 2007 to 2011, where new features as well as new species data were added. However, there have been no new release notes since March 2011.[39]
Compatibility with other sites
MicrobesOnline is compatible with other similar platforms of integrated microbe data, such as IMG and RegTransBase, given that standard identifiers of genes are maintained throughout the database.[40]
MicrobesOnline in the realm of microbe analysis platforms
There have been other efforts to create a unified platform for prokaryote analysis tools, however, most of them focus on one set of analysis types. A few examples of these focused databases include those with an emphasis on metabolic data analysis (Microme [41] ), comparative genomics (MBGD [42] and the OMA Browser [43]), regulons and transcription factors (RegPrecise [44]), comparative functional genomics (Pathline [45]), among many others. However, notable efforts have been made by other teams to create comprehensive platforms that largely overlap with the capabilities of MicrobesOnline. MicroScope [46] and the Integrated Microbial Genomes System[47][48] (IMG) are examples of popular and recently updated databases (As of September 2014).
Extension of metagenome analysis: metaMicrobesOnline
metaMicrobesOnline [49] was compiled by the same developers as MicrobesOnline, and constitutes an extension of MicrobesOnline capacities, by focusing on the phylogenetic analysis of metagenomes. With a similar web interface to MicrobesOnline, the user is capable of toggling between sites via the “switch to” link on the homepage.
See also
- Integrated Microbial Genomes System (IMG)
- BLAST
- BLAT (bioinformatics)
- FASTA format
- Gene ontology
- Hidden Markov model (HMM)
- Homology (biology)
- KEGG
- Multiple Sequence Alignment
External links
- MicrobesOnline home page
- IMG home page reference: Nucleic Acids Research, 2006, Vol. 34, Database issue D344-D348
- Gene Ontology Consortium (GOC): an international collaboration on developing gene ontology and creating annotations of genetic functions.
- The Open Biological and Biomedical Ontologies: a community-based platform that integrates databases and tools of gene ontology.
- Kyoto Encyclopedia of Genes and Genomes (KEGG): a large collection of databases of genes, genomes, biological pathways, chemicals, drugs and diseases.
- NCBI COGs: resources of phylogenetic studies of proteins encoded in complete genomes.
- GLAMM: the Genome-Linked Application for Metabolic Maps, which is an interactive viewer for metabolic pathways and experiments.
- GLAMM Tutorial: a comprehensive guidance for using GLAMM.
- MEGABLAST Search: NCBI's introduction to the MEGABLAST algorithm.
- MeV: MultiExperiment Viewer: a versatile tool for analysing microarray data.
- Microme: a platform that integrates a database, tools and browsers for studying bacterial metabolism.
- MBGD: Microbial Genome Database for Comparative Analysis
- Virtual Institute for Microbial Stress and Survival (VIMSS): the supporter for an integrated program to study how microbes respond to and survive environmental stresses.
References
- Alm, E. J.; Huang, K. H.; Price, M. N.; Koche, R. P.; Keller, K; Dubchak, I. L.; Arkin, A. P. (2005). "The Microbes Online Web site for comparative genomics". Genome Research. 15 (7): 1015–22. doi:10.1101/gr.3844805. PMC 1172046. PMID 15998914.
- Dehal, P. S.; Joachimiak, M. P.; Price, M. N.; Bates, J. T.; Baumohl, J. K.; Chivian, D.; Friedland, G. D.; Huang, K. H.; Keller, K.; Novichkov, P. S.; Dubchak, I. L.; Alm, E. J.; Arkin, A. P. (2009). "Microbes Online: An integrated portal for comparative and functional genomics". Nucleic Acids Research. 38 (Database issue): D396–400. doi:10.1093/nar/gkp919. PMC 2808868. PMID 19906701.
- Feist, A. M.; Herrgård, M. J.; Thiele, I.; Reed, J. L.; Palsson, B. Ø. (2008). "Reconstruction of biochemical networks in microorganisms". Nature Reviews Microbiology. 7 (2): 129–43. doi:10.1038/nrmicro1949. PMC 3119670. PMID 19116616.
- Chen, I. M. A.; Markowitz, V. M.; Chu, K.; Anderson, I.; Mavromatis, K.; Kyrpides, N. C.; Ivanova, N. N. (2013). "Improving Microbial Genome Annotations in an Integrated Database Context". PLoS ONE. 8 (2): e54859. Bibcode:2013PLoSO...854859C. doi:10.1371/journal.pone.0054859. PMC 3570495. PMID 23424620.
- "MicrobesOnline homepage". MicrobesOnline. Retrieved 2014-09-09.
- Virtual Institute for Microbial Stress and Survival; Ernest Orlando Lawrence Berkeley National Laboratory (2008). "Site guide & tutorial". MicrobesOnline. 1 Cyclotron Road • Berkeley, CA 94720.CS1 maint: location (link)
- Dehal, P. S.; Joachimiak, M. P.; Price, M. N.; Bates, J. T.; Baumohl, J. K.; Chivian, D.; Friedland, G. D.; Huang, K. H.; Keller, K.; Novichkov, P. S.; Dubchak, I. L.; Alm, E. J.; Arkin, A. P. (2009). "Microbes Online: An integrated portal for comparative and functional genomics". Nucleic Acids Research. 38 (Database issue): D396–400. doi:10.1093/nar/gkp919. PMC 2808868. PMID 19906701.
- Price, M. N.; Dehal, P. S.; Arkin, A. P. (2008). "FastBLAST: Homology Relationships for Millions of Proteins". PLoS ONE. 3 (10): e3589. Bibcode:2008PLoSO...3.3589P. doi:10.1371/journal.pone.0003589. PMC 2571987. PMID 18974889.
- Barrell, D.; Dimmer, E.; Huntley, R. P.; Binns, D.; O'Donovan, C.; Apweiler, R. (2009). "The GOA database in 2009--an integrated Gene Ontology Annotation resource". Nucleic Acids Research. 37 (Database issue): D396–403. doi:10.1093/nar/gkn803. PMC 2686469. PMID 18957448.
- Virtual Institute for Microbial Stress and Survival; Ernest Orlando Lawrence Berkeley National Laboratory (2008). "Site guide & tutorial". MicrobesOnline. 1 Cyclotron Road • Berkeley, CA 94720.CS1 maint: location (link)
- Kanehisa, M. (2004). "The KEGG resource for deciphering the genome". Nucleic Acids Research. 32 (90001): 277D–280. doi:10.1093/nar/gkh063. PMC 308797. PMID 14681412.
- Bates, J. T.; Chivian, D.; Arkin, A. P. (2011). "GLAMM: Genome-Linked Application for Metabolic Maps". Nucleic Acids Research. 39 (Web Server issue): W400–5. doi:10.1093/nar/gkr433. PMC 3125797. PMID 21624891.
- Virtual Institute for Microbial Stress and Survival; Ernest Orlando Lawrence Berkeley National Laboratory (2008). "Site guide & tutorial". MicrobesOnline. 1 Cyclotron Road • Berkeley, CA 94720.CS1 maint: location (link)
- Alm, E. J.; Huang, K. H.; Price, M. N.; Koche, R. P.; Keller, K; Dubchak, I. L.; Arkin, A. P. (2005). "The Microbes Online Web site for comparative genomics". Genome Research. 15 (7): 1015–22. doi:10.1101/gr.3844805. PMC 1172046. PMID 15998914.
- Pruitt, K. D.; Tatusova, T.; Maglott, D. R. (2007). "NCBI reference sequences (Ref Seq): A curated non-redundant sequence database of genomes, transcripts and proteins". Nucleic Acids Research. 35 (Database issue): D61–5. doi:10.1093/nar/gkl842. PMC 1716718. PMID 17130148.
- Uniprot, Consortium (2009). "The Universal Protein Resource (Uni Prot) 2009". Nucleic Acids Research. 37 (Database issue): D169–74. doi:10.1093/nar/gkn664. PMC 2686606. PMID 18836194.
- Sayers, E. W.; Barrett, T.; Benson, D. A.; Bryant, S. H.; Canese, K.; Chetvernin, V.; Church, D. M.; Dicuccio, M.; Edgar, R.; Federhen, S.; Feolo, M.; Geer, L. Y.; Helmberg, W.; Kapustin, Y.; Landsman, D.; Lipman, D. J.; Madden, T. L.; Maglott, D. R.; Miller, V.; Mizrachi, I.; Ostell, J.; Pruitt, K. D.; Schuler, G. D.; Sequeira, E.; Sherry, S. T.; Shumway, M.; Sirotkin, K.; Souvorov, A.; Starchenko, G.; et al. (2009). "Database resources of the National Center for Biotechnology Information". Nucleic Acids Research. 37 (Database issue): D5–15. doi:10.1093/nar/gkn741. PMC 2686545. PMID 18940862.
- Badger, J. H.; Olsen, G. J. (1999). "CRITICA: Coding region identification tool invoking comparative analysis". Molecular Biology and Evolution. 16 (4): 512–24. doi:10.1093/oxfordjournals.molbev.a026133. PMID 10331277.
- Delcher, A. L.; Bratke, K. A.; Powers, E. C.; Salzberg, S. L. (2007). "Identifying bacterial genes and endosymbiont DNA with Glimmer". Bioinformatics. 23 (6): 673–679. doi:10.1093/bioinformatics/btm009. PMC 2387122. PMID 17237039.
- Wu, C. H.; Nikolskaya, A.; Huang, H.; Yeh, L. S.; Natale, D. A.; Vinayaka, C. R.; Hu, Z. Z.; Mazumder, R.; Kumar, S.; Kourtesis, P.; Ledley, R. S.; Suzek, B. E.; Arminski, L.; Chen, Y.; Zhang, J.; Cardenas, J. L.; Chung, S.; Castro-Alvear, J.; Dinkov, G.; Barker, W. C. (2004). "PIRSF: Family classification system at the Protein Information Resource". Nucleic Acids Research. 32 (90001): 112D–1114. doi:10.1093/nar/gkh097. PMC 308831. PMID 14681371.
- Finn, R. D.; Tate, J.; Mistry, J.; Coggill, P. C.; Sammut, S. J.; Hotz, H. -R.; Ceric, G.; Forslund, K.; Eddy, S. R.; Sonnhammer, E. L. L.; Bateman, A. (2007). "The Pfam protein families database". Nucleic Acids Research. 36 (Database issue): D281–8. doi:10.1093/nar/gkm960. PMC 2238907. PMID 18039703.
- Letunic, I.; Doerks, T.; Bork, P. (2009). "SMART 6: Recent updates and new developments". Nucleic Acids Research. 37 (Database issue): D229–32. doi:10.1093/nar/gkn808. PMC 2686533. PMID 18978020.
- Wilson, D.; Madera, M.; Vogel, C.; Chothia, C.; Gough, J. (2007). "The SUPERFAMILY database in 2007: Families and functions". Nucleic Acids Research. 35 (Database issue): D308–D313. doi:10.1093/nar/gkl910. PMC 1669749. PMID 17098927.
- Tatusov, R. L.; Fedorova, N. D.; Jackson, J. D.; Jacobs, A. R.; Kiryutin, B.; Koonin, E. V.; Krylov, D. M.; Mazumder, R.; Mekhedov, S. L.; Nikolskaya, A. N.; Rao, B. S.; Smirnov, S.; Sverdlov, A. V.; Vasudevan, S.; Wolf, Y. I.; Yin, J. J.; Natale, D. A. (2003). "The COG database: An updated version includes eukaryotes". BMC Bioinformatics. 4: 41. doi:10.1186/1471-2105-4-41. PMC 222959. PMID 12969510.
- Barrell, D.; Dimmer, E.; Huntley, R. P.; Binns, D.; O'Donovan, C.; Apweiler, R. (2009). "The GOA database in 2009--an integrated Gene Ontology Annotation resource". Nucleic Acids Research. 37 (Database issue): D396–403. doi:10.1093/nar/gkn803. PMC 2686469. PMID 18957448.
- Kanehisa, M. (2004). "The KEGG resource for deciphering the genome". Nucleic Acids Research. 32 (90001): 277D–280. doi:10.1093/nar/gkh063. PMC 308797. PMID 14681412.
- Mi, H.; Guo, N.; Kejariwal, A.; Thomas, P. D. (2007). "PANTHER version 6: Protein sequence and function evolution data with expanded representation of biological pathways". Nucleic Acids Research. 35 (Database issue): D247–52. doi:10.1093/nar/gkl869. PMC 1716723. PMID 17130144.
- Mi, H.; Thomas, P. (2009). "PANTHER Pathway: An Ontology-Based Pathway Database Coupled with Data Analysis Tools". Protein Networks and Pathway Analysis. Methods in Molecular Biology. 563. pp. 123–40. doi:10.1007/978-1-60761-175-2_7. ISBN 978-1-60761-174-5. PMID 19597783.
- Selengut, J. D.; Haft, D. H.; Davidsen, T.; Ganapathy, A.; Gwinn-Giglio, M.; Nelson, W. C.; Richter, A. R.; White, O. (2007). "TIGRFAMs and Genome Properties: Tools for the assignment of molecular function and biological process in prokaryotic genomes". Nucleic Acids Research. 35 (Database issue): D260–4. doi:10.1093/nar/gkl1043. PMC 1781115. PMID 17151080.
- Yeats, C.; Lees, J.; Reid, A.; Kellam, P.; Martin, N.; Liu, X.; Orengo, C. (2007). "Gene3D: Comprehensive structural and functional annotation of genomes". Nucleic Acids Research. 36 (Database issue): D414–8. doi:10.1093/nar/gkm1019. PMC 2238970. PMID 18032434.
- Barrett, T.; Troup, D. B.; Wilhite, S. E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I. F.; Soboleva, A.; Tomashevsky, M.; Marshall, K. A.; Phillippy, K. H.; Sherman, P. M.; Muertter, R. N.; Edgar, R. (2009). "NCBI GEO: Archive for high-throughput functional genomic data". Nucleic Acids Research. 37 (Database issue): D885–90. doi:10.1093/nar/gkn764. PMC 2686538. PMID 18940857.
- Faith, J. J.; Driscoll, M. E.; Fusaro, V. A.; Cosgrove, E. J.; Hayete, B.; Juhn, F. S.; Schneider, S. J.; Gardner, T. S. (2007). "Many Microbe Microarrays Database: Uniformly normalized Affymetrix compendia with structured experimental metadata". Nucleic Acids Research. 36 (Database issue): D866–70. doi:10.1093/nar/gkm815. PMC 2238822. PMID 17932051.
- Marraffini, L. A.; Sontheimer, E. J. (2010). "CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea". Nature Reviews Genetics. 11 (3): 181–190. doi:10.1038/nrg2749. PMC 2928866. PMID 20125085.
- Marraffini, L. A.; Sontheimer, E. J. (2010). "CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea". Nature Reviews Genetics. 11 (3): 181–190. doi:10.1038/nrg2749. PMC 2928866. PMID 20125085.
- Bland, C; Ramsey, T. L.; Sabree, F; Lowe, M; Brown, K; Kyrpides, N. C.; Hugenholtz, P (2007). "CRISPR recognition tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats". BMC Bioinformatics. 8: 209. doi:10.1186/1471-2105-8-209. PMC 1924867. PMID 17577412.
- Edgar, R. C. (2007). "PILER-CR: Fast and accurate identification of CRISPR repeats". BMC Bioinformatics. 8: 18. doi:10.1186/1471-2105-8-18. PMC 1790904. PMID 17239253.
- Lowe, T. M.; Eddy, S. R. (1997). "TRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence". Nucleic Acids Research. 25 (5): 955–64. doi:10.1093/nar/25.5.955. PMC 146525. PMID 9023104.
- "MicrobesOnline homepage". MicrobesOnline. Retrieved 2014-09-09.
- "MicrobesOnline release notes". MicrobesOnline. Retrieved 2014-09-10.
- Dehal, P. S.; Joachimiak, M. P.; Price, M. N.; Bates, J. T.; Baumohl, J. K.; Chivian, D.; Friedland, G. D.; Huang, K. H.; Keller, K.; Novichkov, P. S.; Dubchak, I. L.; Alm, E. J.; Arkin, A. P. (2009). "Microbes Online: An integrated portal for comparative and functional genomics". Nucleic Acids Research. 38 (Database issue): D396–400. doi:10.1093/nar/gkp919. PMC 2808868. PMID 19906701.
- "Microme". Microme. Retrieved 2014-09-09.
- Uchiyama, I.; Mihara, M.; Nishide, H.; Chiba, H. (2012). "MBGD update 2013: The microbial genome database for exploring the diversity of microbial world". Nucleic Acids Research. 41 (Database issue): D631–5. doi:10.1093/nar/gks1006. PMC 3531178. PMID 23118485.
- Altenhoff, A. M.; Schneider, A.; Gonnet, G. H.; Dessimoz, C. (2010). "OMA 2011: Orthology inference among 1000 complete genomes". Nucleic Acids Research. 39 (Database issue): D289–94. doi:10.1093/nar/gkq1238. PMC 3013747. PMID 21113020.
- Novichkov, P. S.; Kazakov, A. E.; Ravcheev, D. A.; Leyn, S. A.; Kovaleva, G. Y.; Sutormin, R. A.; Kazanov, M. D.; Riehl, W.; Arkin, A. P.; Dubchak, I.; Rodionov, D. A. (2013). "Reg Precise 3.0 – A resource for genome-scale exploration of transcriptional regulation in bacteria". BMC Genomics. 14: 745. doi:10.1186/1471-2164-14-745. PMC 3840689. PMID 24175918.
- Meyer, M.; Wong, B.; Styczynski, M.; Munzner, T.; Pfister, H. (2010). "Pathline: A Tool for Comparative Functional Genomics". Computer Graphics Forum. 29 (3): 1043–1052. doi:10.1111/j.1467-8659.2009.01710.x.
- Vallenet, D.; Belda, E.; Calteau, A.; Cruveiller, S.; Engelen, S.; Lajus, A.; Le Fevre, F.; Longin, C.; Mornico, D.; Roche, D.; Rouy, Z.; Salvignol, G.; Scarpelli, C.; Thil Smith, A. A.; Weiman, M.; Medigue, C. (2012). "Micro Scope--an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data". Nucleic Acids Research. 41 (Database issue): D636–47. doi:10.1093/nar/gks1194. PMC 3531135. PMID 23193269.
- Markowitz, V. M.; Szeto, E.; Palaniappan, K.; Grechkin, Y.; Chu, K.; Chen, I. M. A.; Dubchak, I.; Anderson, I.; Lykidis, A.; Mavromatis, K.; Ivanova, N. N.; Kyrpides, N. C. (2007). "The integrated microbial genomes (IMG) system in 2007: Data content and analysis tool extensions". Nucleic Acids Research. 36 (Database issue): D528–33. doi:10.1093/nar/gkm846. PMC 2238897. PMID 17933782.
- Markowitz, V. M.; Chen, I. -M. A.; Palaniappan, K.; Chu, K.; Szeto, E.; Pillay, M.; Ratner, A.; Huang, J.; Woyke, T.; Huntemann, M.; Anderson, I.; Billis, K.; Varghese, N.; Mavromatis, K.; Pati, A.; Ivanova, N. N.; Kyrpides, N. C. (2013). "IMG 4 version of the integrated microbial genomes comparative analysis system". Nucleic Acids Research. 42 (Database issue): D560–7. doi:10.1093/nar/gkt963. PMC 3965111. PMID 24165883.
- Chivian, D.; Dehal, P. S.; Keller, K.; Arkin, A. P. (2012). "Meta Microbes Online: Phylogenomic analysis of microbial communities". Nucleic Acids Research. 41 (Database issue): D648–54. doi:10.1093/nar/gks1202. PMC 3531168. PMID 23203984.