Asparagine endopeptidase
Asparagine endopeptidase (AEP, mammalian legumain, δ-secretase; EC 3.4.22.34) is a proteolytic enzyme from C13 peptidase family which hydrolyses a peptide bond using the thiol group of a cysteine residue as a nucleophile (hence also called cysteine protease). It is also known as asparaginyl endopeptidase, citvac, proteinase B, hemoglobinase, PRSC1 gene product or LGMN (Homo sapiens), vicilin peptidohydrolase and bean endopeptidase. In humans it is encoded by the LGMN gene (previous symbol PRSC1).[1][2][3]
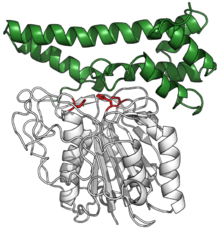
Human pro-legumain with catalytic triad in red, bound to its auto-inhibitory C-terminal prodomain in green. (PDB: 4FGU) | |||||||||
| Identifiers | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| EC number | 3.4.22.34 | ||||||||
| CAS number | 149371-18-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
It hydrolyzes substrates at the C-terminus of asparagine residues. Discovered in 1996 in beans, its homologues have been identified in plants, protozoa, vertebrates, and helminths. The enzyme has been implicated in several human diseases such as cancer, atherosclerosis and inflammation .[4] It can be detected in spleen, liver, brain, testis tissue and heart[5] and the protein is mostly localised to lysosomes and endosomes. It is also interesting that AEP is activated in age-dependent manner.[6]
Distribution
This enzyme was originally identified in the vacuoles of legume seeds, and was subsequently identified the lysosomes of mammals and Schistosoma mansoni.[7] They are now known to be present in a range of plants and animals.[8][9]
Activity
Reaction and specificity
This enzyme catalyses the following chemical reaction:
- Hydrolysis of proteins and small molecule substrates at -Asn-Xaa- bonds
Both plant and animal legumains are most active in acidic environments.[10][11]
Prodomain processing
Legmains are produced as inactive precursor zymogens. their C-terminal domain binds over their active site (where a substrate would normally bind), inhibiting activity.[10] Once in the acidic environment of the vacuole or lysosome, the prodomain is cleaved off to reveal the active enzyme.[10][11]
Mechanism
Legumain is a cysteine protease from the C13 family of the CD clan of proteases (MEROPS).[11] It uses a catalytic triad of Cysteine-Histidine-Asparagine in its active site to perform covalent proteolysis of its substrate.[3]
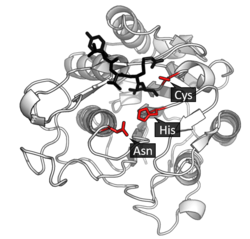
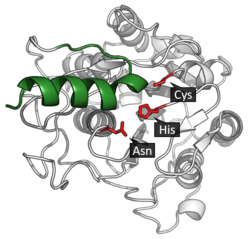
Activation
Asparagine endopeptidase is synthesized as an inactive zymogen.[12] AEP and other cysteine peptidase are activated when pH changes from neutral to acidic. It undergoes autoproteolytic maturation for catalytic activation. It appears to be autocatalytically cleaved after asparagine or aspartate residue. Activation begins at pH 4.5. The chemical structure at this point shows that breaks which occurs at pH 4.5 can be healed under the basic crystallization conditions. C-terminal fragments (∼13 kDa) generated during autoproteolysis can gradually re-ligated to form the proenzyme when the pH is increased towards pH 7.5, which means that proteolytic activation of AEP can be reversible.[6]
Biological roles
Plants
Antimicrobial activity
In plant Oldenlandia affinis it generates antimicrobial cyclic peptides which are important for defence against pathogens in plants.[13][14] The herb has been used in native African medicine to accelerate childbirth.[13]
Animals
Innate immune system
There are many regulators which affect immune system and help to keep it balanced. If the immune system is too active there is a danger of developing an autoimmune disease, while passive immune system will lead to infections or cancer. Antigen presenting is a key role in activation of immune system.[4] It has been discovered that AEP plays role in this critical moment. AEP is involved is presenting of foreign and self proteins using MHCII protein complex.[15] The role of AEP in immunity is not clear, but it seems that it is connected with checkpoint inhibitors such as PD-1, which downregulates AEP which is key to shifting the balance between cancer fighting cells and regulatory T cells. In the absence of AEP, inhibitory checkpoints may not have a beneficial response. Measuring of this enzyme in patients could predict which one of them may provide better response to treatment.[16]
Signalling
In innate immunity TLRs play an important role. These TLRs (mainly TLR7 and TLR9) can be proteolytically activated by AEP.[17] The reduction of proinflammatory cytokines by stimulating TLR9 was found in myeloid cells and plasmacytoid dendritic cells which lacked AEP.[18] Enzyme is also important in processing of influenza virus and immune response using TLR7.[17] AEP plays a critical role in TLR processing. and AEP can initiate removal of invariant chain in MHC-II complex, which can critically influence peptide generation and activity of MHCII.[19]
Disease
Neurodegenerative diseases
AEP is activated during brain ischemia or brain acidosis and epilepsia seizure. It digests SET protein, which is an inhibitor of DNase, leading to DNA damage and causing damage of the brain. Increased activity of AEP in brain is also observed in patients with Alzheimer's disease and Parkinson's disease (PD). AEP cleaves tau protein and amyloid precursor protein. In patients with PD, alpha synuclein is cut by AEP into toxic chunks.[4]
Alzheimer's disease
Active AEP was found at increased levels and translocated to the cytoplasm of neuronal cells of AD patients.[20] In AD the plaques are composed of amyloid beta, intracellular neurofibrillary tangles and tau protein. The dysfunction of APP proteolysis and the abnormal phosphorylation of tau lead to the formation of neuritic plaques and neurofibrillary tangles (NFTs), respectively, causing neuronal degeneration and dementia[21] It also play a crucial role in behavior disorders connected with AD such as anxiety and depression. It also plays a role in stroke. Since stroke elicits acidity in the brain AEP become active due to low pH level. Then it cleaves SET which causes death of brain cells.[22] Targeting of AEP might help to prevent onset of AD symptoms. Development of AEP-selective inhibitors (such as Cbz-L-Ala-L-Ala-AzaAsnchloromethylketone and aza-peptidyl AEP inhibitors) is crucial for helping with diseases.[4]
References
- Hara-Nishimura I (1998). "Asparaginyl endopeptidase". In Barrett AJ, Rawlings ND, Woessner JF (eds.). Handbook of Proteolytic Enzymes. London: Academic Press. pp. 746–749.
- Dalton JP, Brindley PJ (1998). "Schistosome legumain". In Barrett AJ, Rawlings ND, Woessner JF (eds.). Handbook of Proteolytic Enzymes. London: Academic Press. pp. 749–754.
- Chen JM, Rawlings ND, Stevens RA, Barrett AJ (December 1998). "Identification of the active site of legumain links it to caspases, clostripain and gingipains in a new clan of cysteine endopeptidases". FEBS Letters. 441 (3): 361–5. doi:10.1016/S0014-5793(98)01574-9. PMID 9891971.
- Zhang Z, Xie M, Ye K (October 2016). "Asparagine endopeptidase is an innovative therapeutic target for neurodegenerative diseases". Expert Opinion on Therapeutic Targets. 20 (10): 1237–45. doi:10.1080/14728222.2016.1182990. PMC 5315368. PMID 27115710.
- Chen JM, Dando PM, Stevens RA, Fortunato M, Barrett AJ (October 1998). "Cloning and expression of mouse legumain, a lysosomal endopeptidase". The Biochemical Journal. 335 ( Pt 1) (1): 111–7. doi:10.1042/bj3350111. PMC 1219758. PMID 9742219.
- Zhao L, Hua T, Crowley C, Ru H, Ni X, Shaw N, Jiao L, Ding W, Qu L, Hung LW, Huang W, Liu L, Ye K, Ouyang S, Cheng G, Liu ZJ (March 2014). "Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage". Cell Research. 24 (3): 344–58. doi:10.1038/cr.2014.4. PMC 3945893. PMID 24407422.
- Chen JM, Dando PM, Rawlings ND, Brown MA, Young NE, Stevens RA, Hewitt E, Watts C, Barrett AJ (March 1997). "Cloning, isolation, and characterization of mammalian legumain, an asparaginyl endopeptidase". The Journal of Biological Chemistry. 272 (12): 8090–8. doi:10.1074/jbc.272.12.8090. PMID 9065484.
- Müntz K, Shutov AD (August 2002). "Legumains and their functions in plants". Trends in Plant Science. 7 (8): 340–4. doi:10.1016/S1360-1385(02)02298-7. PMID 12167328.
- Shutov AD, Blattner FR, Kakhovskaya IA, Müntz K (February 2012). "New aspects of the molecular evolution of legumains, Asn-specific cysteine proteinases". Journal of Plant Physiology. 169 (3): 319–21. doi:10.1016/j.jplph.2011.11.005. PMID 22196948.
- Chen JM, Fortunato M, Barrett AJ (December 2000). "Activation of human prolegumain by cleavage at a C-terminal asparagine residue". The Biochemical Journal. 352 Pt 2 (2): 327–34. doi:10.1042/bj3520327. PMC 1221463. PMID 11085925.
- "C13 family". merops.sanger.ac.uk. MEROPS - the Peptidase Database. Retrieved 2016-06-09.
- Li DN, Matthews SP, Antoniou AN, Mazzeo D, Watts C (October 2003). "Multistep autoactivation of asparaginyl endopeptidase in vitro and in vivo". The Journal of Biological Chemistry. 278 (40): 38980–90. doi:10.1074/jbc.m305930200. PMID 12860980.
- Gillon AD, Saska I, Jennings CV, Guarino RF, Craik DJ, Anderson MA (February 2008). "Biosynthesis of circular proteins in plants". The Plant Journal. 53 (3): 505–15. doi:10.1111/j.1365-313x.2007.03357.x. PMID 18086282.
- Craik DJ (February 2012). "Host-defense activities of cyclotides". Toxins. 4 (2): 139–56. doi:10.3390/toxins4020139. PMC 3317112. PMID 22474571.
- Matthews SP, Werber I, Deussing J, Peters C, Reinheckel T, Watts C (March 2010). "Distinct protease requirements for antigen presentation in vitro and in vivo". Journal of Immunology. 184 (5): 2423–31. doi:10.4049/jimmunol.0901486. PMID 20164435.
- "Enzyme AEP's importance to immunity discovered". Retrieved 2018-08-28.
- Maschalidi S, Hässler S, Blanc F, Sepulveda FE, Tohme M, Chignard M, van Endert P, Si-Tahar M, Descamps D, Manoury B (2012-08-16). "Asparagine endopeptidase controls anti-influenza virus immune responses through TLR7 activation". PLoS Pathogens. 8 (8): e1002841. doi:10.1371/journal.ppat.1002841. PMC 3420946. PMID 22916010.
- Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E, Lennon-Duménil AM, Amigorena S, Cabanie L, Manoury B (November 2009). "Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells". Immunity. 31 (5): 737–48. doi:10.1016/j.immuni.2009.09.013. PMID 19879164.
- Manoury B, Mazzeo D, Li DN, Billson J, Loak K, Benaroch P, Watts C (April 2003). "Asparagine endopeptidase can initiate the removal of the MHC class II invariant chain chaperone". Immunity. 18 (4): 489–98. doi:10.1016/s1074-7613(03)00085-2. PMID 12705852.
- Basurto-Islas G, Grundke-Iqbal I, Tung YC, Liu F, Iqbal K (June 2013). "Activation of asparaginyl endopeptidase leads to Tau hyperphosphorylation in Alzheimer disease". The Journal of Biological Chemistry. 288 (24): 17495–507. doi:10.1074/jbc.M112.446070. PMC 3682549. PMID 23640887.
- Zhang Z, Song M, Liu X, Kang SS, Kwon IS, Duong DM, Seyfried NT, Hu WT, Liu Z, Wang JZ, Cheng L, Sun YE, Yu SP, Levey AI, Ye K (November 2014). "Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer's disease". Nature Medicine. 20 (11): 1254–62. doi:10.1038/nm.3700. PMC 4224595. PMID 25326800.
- Gao J, Li K, Du L, Yin H, Tan X, Yang Z (July 2018). "Deletion of asparagine endopeptidase reduces anxiety- and depressive-like behaviors and improves abilities of spatial cognition in mice". Brain Research Bulletin. 142: 147–155. doi:10.1016/j.brainresbull.2018.07.010. PMID 30030107.
External links
- Legumain at the US National Library of Medicine Medical Subject Headings (MeSH)