3-hydroxyisobutyrate dehydrogenase
In enzymology, a 3-hydroxyisobutyrate dehydrogenase (EC 1.1.1.31) also known as β-hydroxyisobutyrate dehydrogenase or 3-hydroxyisobutyrate dehydrogenase, mitochondrial (HIBADH) is an enzyme[5] that in humans is encoded by the HIBADH gene.[6]
| 3-hydroxyisobutyrate dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.1.1.31 | ||||||||
| CAS number | 9028-39-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
3-Hydroxyisobutyrate dehydrogenase catalyzes the chemical reaction:
- 3-hydroxy-2-methylpropanoate + NAD+ 2-methyl-3-oxopropanoate + NADH + H+
Thus, the two substrates of this enzyme are 3-hydroxy-2-methylpropanoate and NAD+, whereas its 3 products are 2-methyl-3-oxopropanoate, NADH, and H+.
This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is 3-hydroxy-2-methylpropanoate:NAD+ oxidoreductase. This enzyme participates in valine, leucine and isoleucine degradation.
Function
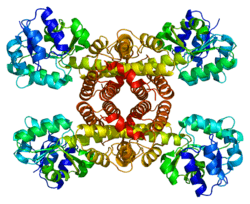
3-hydroxyisobutyrate dehydrogenase is a tetrameric mitochondrial enzyme that catalyzes the NAD+-dependent, reversible oxidation of 3-hydroxyisobutyrate, an intermediate of valine catabolism, to methylmalonate semialdehyde.[6]
Structural studies
As of late 2007, five structures have been solved for this class of enzymes, with PDB accession codes 1WP4, 2CVZ, 2GF2, 2H78, and 2I9P.
References
- GRCh38: Ensembl release 89: ENSG00000106049 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000029776 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Robinson WG, Coon MJ (March 1957). "The purification and properties of beta-hydroxyisobutyric dehydrogenase". J. Biol. Chem. 225 (1): 511–21. PMID 13416257.
- "Entrez Gene: HIBADH 3-hydroxyisobutyrate dehydrogenase".
Further reading
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Hillier LW, Fulton RS, Fulton LA, et al. (2003). "The DNA sequence of human chromosome 7". Nature. 424 (6945): 157–64. Bibcode:2003Natur.424..157H. doi:10.1038/nature01782. PMID 12853948.
- Scherer SW, Cheung J, MacDonald JR, et al. (2003). "Human chromosome 7: DNA sequence and biology". Science. 300 (5620): 767–72. Bibcode:2003Sci...300..767S. doi:10.1126/science.1083423. PMC 2882961. PMID 12690205.
- Mammalian Gene Collection Program Team; Strausberg, R. L.; Feingold, E. A.; Grouse, L. H.; et al. (2002). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proceedings of the National Academy of Sciences. 99 (26): 16899–16903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Sanger Centre, The; Washington University Genome Sequencing Cente, The (1999). "Toward a complete human genome sequence". Genome Res. 8 (11): 1097–108. doi:10.1101/gr.8.11.1097. PMID 9847074.
- Hughes GJ, Frutiger S, Paquet N, et al. (1994). "Human liver protein map: update 1993". Electrophoresis. 14 (11): 1216–22. doi:10.1002/elps.11501401181. PMID 8313870.
- Rougraff PM, Paxton R, Kuntz MJ, et al. (1988). "Purification and characterization of 3-hydroxyisobutyrate dehydrogenase from rabbit liver". J. Biol. Chem. 263 (1): 327–31. PMID 3335502.
- Rougraff PM, Zhang B, Kuntz MJ, et al. (1989). "Cloning and sequence analysis of a cDNA for 3-hydroxyisobutyrate dehydrogenase. Evidence for its evolutionary relationship to other pyridine nucleotide-dependent dehydrogenases". J. Biol. Chem. 264 (10): 5899–903. PMID 2647728.
External links
- Human HIBADH genome location and HIBADH gene details page in the UCSC Genome Browser.
- PDBe-KB provides an overview of all the structure information available in the PDB for Human 3-hydroxyisobutyrate dehydrogenase, mitochondrial