Mannose
Mannose, packaged as the nutritional supplement "d-mannose", is a sugar monomer of the aldohexose series of carbohydrates. It is a C-2 epimer of glucose. Mannose is important in human metabolism, especially in the glycosylation of certain proteins. Several congenital disorders of glycosylation are associated with mutations in enzymes involved in mannose metabolism.[1]
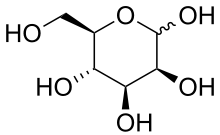 D-Mannopyranose | |
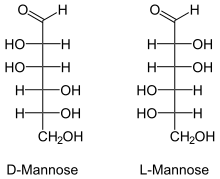 Fischer projections | |
| Identifiers | |
|---|---|
| ChEMBL | |
| ChemSpider |
|
| ECHA InfoCard | 100.007.705 |
| MeSH | Mannose |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| Properties | |
| C6H12O6 | |
| Molar mass | 180.156 g·mol−1 |
| -102.90·10−6 cm3/mol | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Mannose is not an essential nutrient; it can be produced in the human body from glucose, or converted into glucose. Mannose provides 2-5 kcal/g. It is partially excreted in the urine.
Structure
Mannose commonly exists as two different-sized rings, the pyranose (six-membered) form and the furanose (five-membered) form. Each ring closure can have either an alpha or beta configuration at the anomeric position. The chemical rapidly undergoes isomerization among these four forms.
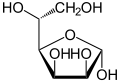 α-D-Mannofuranose | |
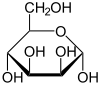 α-D-Mannopyranose 67% |
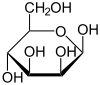 β-D-Mannopyranose 33% |
Metabolism
While much of the mannose used in glycosylation is believed to be derived from glucose, in cultured hepatoma cells (cancerous cells from the liver), most of the mannose for glycoprotein biosynthesis comes from extracellular mannose, not glucose.[2] Many of the glycoproteins produced in the liver are secreted into the bloodstream, so dietary mannose is distributed throughout the body. [3]
Mannose is present in numerous glycoconjugates including N-linked glycosylation of proteins. C-Mannosylation is also abundant and can be found in collagen-like regions.
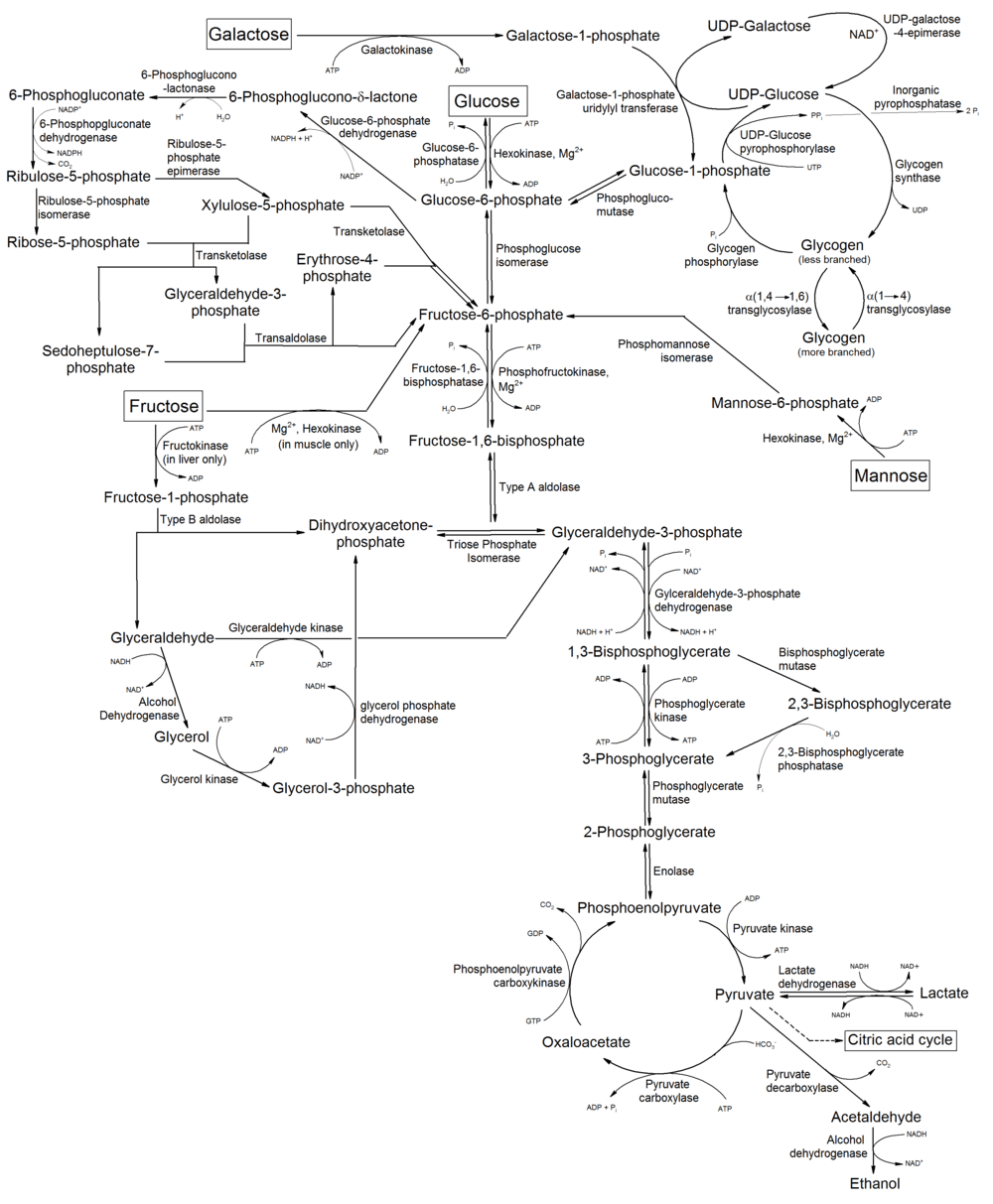
The digestion of many polysaccharides and glycoproteins yields mannose, which is phosphorylated by hexokinase to generate mannose-6-phosphate. Mannose-6-phosphate is converted to fructose-6-phosphate, by the enzyme phosphomannose isomerase, and then enters the glycolytic pathway or is converted to glucose-6-phosphate by the gluconeogenic pathway of hepatocytes.
Mannose is a dominant monosaccharide in N-linked glycosylation, which is a post-translational modification of proteins. It is initiated by the en bloc transfer on Glc3Man9GlcNAc2 to nascent glycoproteins in the endoplasmic reticulum in a co-translational manner as the protein entered through the transport system. Glucose is hydrolyzed on fully folded protein and the mannose moieties are hydrolyzed by ER and Golgi-resident mannosidases. Typically, mature human glycoproteins only contain three mannose residues buried under sequential modification by GlcNAc, galactose, and sialic acid. This is important, as the innate immune system in mammals is geared to recognise exposed mannose residues. This activity is due to the prevalence of mannose residues, in the form of mannans, on the surfaces of yeasts. The human immunodeficiency virus displays considerable amount of mannose residues due to the tight clustering of glycans in its viral spike.[4][5] These mannose residues are the target for broadly neutralizing antibodies.[6]
Biotechnology
Recombinant proteins produced in yeast may be subject to mannose addition in patterns different from those used by mammalian cells.[7] This difference in recombinant proteins from those normally produced in mammalian organisms may influence the effectiveness of vaccines.
Formation
Mannose can be formed by the oxidation of mannitol.
It can also be formed from glucose in the Lobry-de Bruyn-van Ekenstein transformation.
Etymology
The root of both "mannose" and "mannitol" is manna, which the Bible records as the food supplied to the Israelites during their journey in the region of Sinai. Several trees and shrubs can produce a substance called manna, such as the "manna tree" (Fraxinus ornus) from whose secretions mannitol was originally isolated.
Uses
Mannose (D-mannose) is used as a nutritional supplement to prevent recurrent urinary tract infections. [8]
Configuration
Mannose differs from glucose by inversion of the C-2 chiral center. Mannose displays a pucker in the solution ring form. This simple change leads to the drastically different biochemistry of the two hexoses. This change has the same effect on the other aldohexoses, as well.
Mannose PTS permease
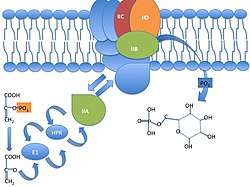
The PEP-dependent sugar transporting phosphotransferase system transports and simultaneously phosphorylates its sugar substrates. Mannose XYZ permease is a member of the family, with this distinct method being used by bacteria for sugar uptake particularly exogenous hexoses in the case of mannose XYZ to release the phosphate esters into the cell cytoplasm in preparation for metabolism primarily through the route of glycolysis.[9] The MANXYZ transporter complex is also involved in infection of E. coli by bacteriophage lambda, with subunit ManY and ManZ being sufficient for proper lambda phage infection.[10] MANXYZ possesses four domains in three polypeptide chains; ManX, ManY, and ManZ. The ManX subunit forms a homodimer that is localized to the cytoplasmic side of the membrane. ManX contains two domains IIA and IIB linked by a hinge peptide with each domain containing a phosphorylation site and phosphoryl transfer occurs between both subunits.[11] ManX can be membrane bound or not.[10] The ManY and ManNZ subunits are hydrophobic integral membrane proteins with six and one transmembrane alpha helical spanner(s).[12] [13]The phosphoryl group of PEP is transferred to the imported sugar via Enzyme 1, histidine protein phosphate carrier, and then to the ManX, ManY, and ManZ subunits of the ManXYZ transportation complex, which phosphorylates the entering hexose sugar, creating a hexose-6-phosphate.
See also
- α-Mannosidase
- Mannose receptor
- Mannan oligosaccharide-based nutritional supplements
- Rhamnose, 6-deoxy-L-mannose
References
- Freeze, H. H.; Sharma, V. (2010). "Metabolic manipulation of glycosylation disorders in humans and animal models". Seminars in Cell & Developmental Biology. 21 (6): 655–662. doi:10.1016/j.semcdb.2010.03.011. PMC 2917643. PMID 20363348.
- Alton, G.; Hasilik, M.; Niehues, R.; Panneerselvam, K.; Etchison, J. R.; Fana, F.; Freeze, H. H. (1998). "Direct utilization of mannose for mammalian glycoprotein biosynthesis". Glycobiology. 8 (3): 285–295. doi:10.1093/glycob/8.3.285. PMID 9451038.
- Davis, J. A.; Freeze, H. H. (2001). "Studies of mannose metabolism and effects of long-term mannose ingestion in the mouse". Biochimica et Biophysica Acta (BBA) - General Subjects. 1528 (2–3): 116–126. doi:10.1016/S0304-4165(01)00183-0. PMID 11687298.
- Pritchard, Laura K.; Spencer, Daniel I. R.; Royle, Louise; Bonomelli, Camille; Seabright, Gemma E.; Behrens, Anna-Janina; Kulp, Daniel W.; Menis, Sergey; Krumm, Stefanie A. (2015-06-24). "Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies". Nature Communications. 6: 7479. Bibcode:2015NatCo...6.7479P. doi:10.1038/ncomms8479. PMC 4500839. PMID 26105115.
- Pritchard, Laura K.; Vasiljevic, Snezana; Ozorowski, Gabriel; Seabright, Gemma E.; Cupo, Albert; Ringe, Rajesh; Kim, Helen J.; Sanders, Rogier W.; Doores, Katie J. (2015-06-16). "Structural Constraints Determine the Glycosylation of HIV-1 Envelope Trimers". Cell Reports. 11 (10): 1604–1613. doi:10.1016/j.celrep.2015.05.017. ISSN 2211-1247. PMC 4555872. PMID 26051934.
- Crispin, Max; Doores, Katie J (2015-04-01). "Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design". Current Opinion in Virology. Viral pathogenesis • Preventive and therapeutic vaccines. 11: 63–69. doi:10.1016/j.coviro.2015.02.002. PMC 4827424. PMID 25747313.
- Vlahopoulos, S.; Gritzapis, A. D.; Perez, S. A.; Cacoullos, N.; Papamichail, M.; Baxevanis, C. N. (2009). "Mannose addition by yeast Pichia pastoris on recombinant HER-2 protein inhibits recognition by the monoclonal antibody herceptin". Vaccine. 27 (34): 4704–4708. doi:10.1016/j.vaccine.2009.05.063. PMID 19520203.
- Porru, D.; Parmigiani, A.; Tinelli, C.; Barletta, D.; Choussos, D.; Di Franco, C.; Rovereto, B. (2014). "Oral D-mannose in recurrent urinary tract infections in women: A pilot study". Journal of Clinical Urology. 7 (3): 208–213. doi:10.1177/2051415813518332.
- Postma, P. W.; Lengeler, J. W.; Jacobson, G. R. (1993). "Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria". Microbiological Reviews. 57 (3): 543–594. doi:10.1128/MMBR.57.3.543-594.1993. PMC 372926. PMID 8246840.
- Erni, B.; Zanolari, B. (1985). "The mannose-permease of the bacterial phosphotransferase system. Gene cloning and purification of the enzyme IIMan/IIIMan complex of Escherichia coli". The Journal of Biological Chemistry. 260 (29): 15495–15503. PMID 2999119.
- Erni, B.; Zanolari, B.; Graff, P.; Kocher, H. P. (1989). "Mannose permease of Escherichia coli. Domain structure and function of the phosphorylating subunit". The Journal of Biological Chemistry. 264 (31): 18733–18741. PMID 2681202.
- Huber, F.; Erni, B. (1996). "Membrane topology of the mannose transporter of Escherichia coli K12". European Journal of Biochemistry / FEBS. 239 (3): 810–817. doi:10.1111/j.1432-1033.1996.0810u.x. PMID 8774730.
- Liu, Xueli; Zeng, Jianwei; Huang, Kai; Wang, Jiawei (2019-06-17). "Structure of the mannose transporter of the bacterial phosphotransferase system". Cell Research. 29 (8): 680–682. doi:10.1038/s41422-019-0194-z. ISSN 1748-7838. PMC 6796895. PMID 31209249.