Transgenerational epigenetic inheritance
Transgenerational epigenetic inheritance, (TEI), is the transmission of epigenetic markers from one organism to the next (i.e., parent–child transmission) that affects the traits of offspring without alteration of the primary structure of DNA (i.e. the sequence of nucleotides)[2]:168[3]—in other words, epigenetically. The less precise term "epigenetic inheritance" may cover both cell–cell and organism–organism information transfer. Although these two levels of epigenetic inheritance are equivalent in unicellular organisms, they may have distinct mechanisms and evolutionary distinctions in multicellular organisms.
The environment can induce the epigenetic marks (epigenetic tags) for some epigenetically influenced traits,[2] and some marks are heritable,[2] leading some to view epigenetics as a relaxation of biology's rejection of the inheritance of acquired characteristics (Lamarckism).[3]
Epigenetic categories
Four general categories of epigenetic modification are known:[4]
- self-sustaining metabolic loops, in which a mRNA or protein product of a gene stimulates transcription of the gene; e.g. Wor1 gene in Candida albicans[5]
- structural templating in which structures are replicated using a template or scaffold structure on the parent; e.g. the orientation and architecture of cytoskeletal structures, cilia and flagella,[6] prions, proteins that replicate by changing the structure of normal proteins to match their own[7]
- chromatin marks, in which methyl or acetyl groups bind to DNA nucleotides or histones thereby altering gene expression patterns; e.g. Lcyc gene in Linaria vulgaris described below
- RNA silencing, in which small RNA strands interfere (RNAi) with the transcription of DNA or translation of mRNA; known only from a few studies, mostly in Caenorhabditis elegans[8]
Inheritance of epigenetic marks
Epigenetic variation may take one of four general forms. Others may yet be elucidated, but currently self-sustaining feedback loops, spatial templating, chromatin marking, and RNA-mediated pathways modify epigenes at the level of individual cells. Epigenetic variation within multicellular organisms may be endogenous, generated by cell–cell signaling (e.g. during cell differentiation early in development), or exogenous, a cellular response to environmental cues.
Removal vs. retention
In sexually reproducing organisms, much of the epigenetic modification within cells is reset during meiosis (e.g. marks at the FLC locus controlling plant vernalization[9]), though some epigenetic responses have been shown to be conserved (e.g. transposon methylation in plants[9]). Differential inheritance of epigenetic marks due to underlying maternal or paternal biases in removal or retention mechanisms may lead to the assignment of epigenetic causation to some parent of origin effects in animals[10] and plants.[11]
Reprogramming
In mammals, epigenetic marks are erased during two phases of the life cycle. Firstly just after fertilization and secondly, in the developing primordial germ cells, the precursors to future gametes.[2] During fertilization the male and female gametes join in different cell cycle states and with different configuration of the genome. The epigenetic marks of the male are rapidly diluted. First, the protamines associated with male DNA are replaced with histones from the female's cytoplasm, most of which are acetylated due to either higher abundance of acetylated histones in the female's cytoplasm or through preferential binding of the male DNA to acetylated histones.[12][13] Second, male DNA is systematically demethylated in many organisms,[14][15] possibly through 5-hydroxymethylcytosine. However, some epigenetic marks, particularly maternal DNA methylation, can escape this reprogramming; leading to parental imprinting.
In the primordial germ cells (PGC) there is a more extensive erasure of epigenetic information. However, some rare sites can also evade erasure of DNA methylation.[16] If epigenetic marks evade erasure during both zygotic and PGC reprogramming events, this could enable transgenerational epigenetic inheritance.
Recognition of the importance of epigenetic programming to the establishment and fixation of cell line identity during early embryogenesis has recently stimulated interest in artificial removal of epigenetic programming.[17] Epigenetic manipulations may allow for restoration of totipotency in stem cells or cells more generally, thus generalizing regenerative medicine.
Retention
Cellular mechanisms may allow for co-transmission of some epigenetic marks. During replication, DNA polymerases working on the leading and lagging strands are coupled by the DNA processivity factor proliferating cell nuclear antigen (PCNA), which has also been implicated in patterning and strand crosstalk that allows for copy fidelity of epigenetic marks.[18][19] Work on histone modification copy fidelity has remained in the model phase, but early efforts suggest that modifications of new histones are patterned on those of the old histones and that new and old histones randomly assort between the two daughter DNA strands.[20] With respect to transfer to the next generation, many marks are removed as described above. Emerging studies are finding patterns of epigenetic conservation across generations. For instance, centromeric satellites resist demethylation.[21] The mechanism responsible for this conservation is not known, though some evidence suggests that methylation of histones may contribute.[21][22] Dysregulation of the promoter methylation timing associated with gene expression dysregulation in the embryo was also identified.[23]
Decay
Whereas the mutation rate in a given 100-base gene may be 10−7 per generation, epigenes may "mutate" several times per generation or may be fixed for many generations.[24] This raises the question: do changes in epigene frequencies constitute evolution? Rapidly decaying epigenetic effects on phenotypes (i.e. lasting less than three generations) may explain some of the residual variation in phenotypes after genotype and environment are accounted for. However, distinguishing these short-term effects from the effects of the maternal environment on early ontogeny remains a challenge.
Contribution to phenotypes
The relative importance of genetic and epigenetic inheritance is subject to debate.[25][26] Though hundreds of examples of epigenetic modification of phenotypes have been published,[27][28] few studies have been conducted outside of the laboratory setting.[29] Therefore, the interactions of genes and epigenes with the environment cannot be inferred despite the central role of environment in natural selection. Experimental methodologies for manipulating epigenetic mechanisms are nascent (e.g.[30]) and will need rigorous demonstration before studies explicitly testing the relative contributions of genotype, environment, and epigenotype are feasible.
In plants

Studies concerning transgenerational epigenetic inheritance in plants have been reported as early as the 1950s.[31] One of the earliest and best characterized examples of this is b1 paramutation in maize.[31][32][33][34][35][36][37][38] The b1 gene encodes a basic helix-loop-helix transcription factor that is involved in the anthocyanin production pathway. When the b1 gene is expressed, the plant accumulates anthocyanin within its tissues, leading to a purple coloration of those tissues. The B-I allele (for B-Intense) has high expression of b1 resulting in the dark pigmentation of the sheath and husk tissues while the B' (pronounced B-prime) allele has low expression of b1 resulting in low pigmentation in those tissues.[39] When homozygous B-I parents are crossed to homozygous B', the resultant F1 offspring all display low pigmentation which is due to gene silencing of b1.[31][39] Unexpectedly, when F1 plants are self-crossed, the resultant F2 generation all display low pigmentation and have low levels of b1 expression. Furthermore, when any F2 plant (including those that are genetically homozygous for B-I) are crossed to homozygous B-I, the offspring will all display low pigmentation and expression of b1.[31][39] The lack of darkly pigmented individuals in the F2 progeny is an example of non-Mendelian inheritance and further research has suggested that the B-I allele is converted to B' via epigenetic mechanisms.[33][34] The B' and B-I alleles are considered to be epialleles because they are identical at the DNA sequence level but differ in the level of DNA methylation, siRNA production, and chromosomal interactions within the nucleus.[37][40][36][35] Additionally, plants defective in components of the RNA-directed DNA-methylation pathway show an increased expression of b1 in B' individuals similar to that of B-I, however, once these components are restored, the plant reverts to the low expression state.[38][41][42][43] Although spontaneous conversion from B-I to B' has been observed, a reversion from B' to B-I (green to purple) has never been observed over 50 years and thousands of plants in both greenhouse and field experiments.[44]
Examples of environmentally induced transgenerational epigenetic inheritance in plants has also been reported.[45][46][47] In one case, rice plants that were exposed to drought-simulation treatments displayed increased tolerance to drought after 11 generations of exposure and propagation by single-seed descent as compared to non-drought treated plants.[45] Differences in drought tolerance was linked to directional changes in DNA-methylation levels throughout the genome, suggesting that stress-induced heritable changes in DNA-methylation patterns may be important in adaptation to recurring stresses.[45] In another study, plants that were exposed to moderate caterpillar herbivory over multiple generations displayed increased resistance to herbivory in subsequent generations (as measured by caterpillar dry mass) compared to plants lacking herbivore pressure.[46] This increase in herbivore resistance persisted after a generation of growth without any herbivore exposure suggesting that the response was transmitted across generations.[46] The report concluded that components of the RNA-directed DNA-methylation pathway are involved in the increased resistance across generations.[46]
In humans
A number of studies suggest the existence of transgenerational epigenetic inheritance in humans.[2] These include those of the Dutch famine of 1944–45, wherein the offspring born during the famine were smaller than those born the year before the famine and the effects could last for two generations. Moreover, these offspring were found to have an increased risk of glucose intolerance in adulthood.[48] Differential DNA methylation has been found in adult female offspring who had been exposed to famine in utero, but it is unknown whether these differences are present in their germline.[48] It is hypothesized that inhibiting the PIM3 gene may have caused slower metabolism in later generations, but causation has not been proven, only correlation.[49] The phenomenon is sometimes referred to as Dutch Hunger Winter Syndrome. Another study hypothesizes epigenetic changes on the Y chromosome to explain differences in lifespan among the male descendants of prisoners of war in the American Civil War.[50]
The Överkalix study noted sex-specific effects; a greater body mass index (BMI) at 9 years in sons, but not daughters, of fathers who began smoking early. The paternal grandfather's food supply was only linked to the mortality RR of grandsons and not granddaughters. The paternal grandmother's food supply was only associated with the granddaughters' mortality risk ratio. When the grandmother had a good food supply was associated with a twofold higher mortality (RR). This transgenerational inheritance was observed with exposure during the slow growth period (SGP). The SGP is the time before the start of puberty, when environmental factors have a larger impact on the body. The ancestors' SGP in this study was set between the ages of 9-12 for boys and 8–10 years for girls. This occurred in the SGP of both grandparents, or during the gestation period/infant life of the grandmothers, but not during either grandparent's puberty. The father's poor food supply and the mother's good food supply were associated with a lower risk of cardiovascular death.[48][51]
The loss of genetic expression which results in Prader–Willi syndrome or Angelman syndrome has in some cases been found to be caused by epigenetic changes (or "epimutations") on both the alleles, rather than involving any genetic mutation. In all 19 informative cases, the epimutations that, together with physiological imprinting and therefore silencing of the other allele, were causing these syndromes were localized on a chromosome with a specific parental and grandparental origin. Specifically, the paternally derived chromosome carried an abnormal maternal mark at the SNURF-SNRPN, and this abnormal mark was inherited from the paternal grandmother.[48]
Similarly, epimutations on the MLH1 gene has been found in two individuals with a phenotype of hereditary nonpolyposis colorectal cancer, and without any frank MLH1 mutation which otherwise causes the disease. The same epimutations were also found on the spermatozoa of one of the individuals, indicating the potential to be transmitted to offspring.[48]
A study has shown childhood abuse (defined in this study as "sexual contact, severe physical abuse and/or severe neglect") leads to epigenetic modifications of glucocorticoid receptor expression[52][53] which play a role in HPA (hypothalamic-pituitary-adrenal) activity. Animal experiments have shown that epigenetic changes depend on mother-infant interactions after birth.[54] In a recent study investigating correlations among maternal stress in pregnancy and methylation in teenagers and their mothers, it has been found that children of women who were abused during pregnancy were significantly more likely than others to have methylated glucocorticoid-receptor genes,[55] which in turn change the response to stress, leading to a higher susceptibility to anxiety.
Effects on fitness
Epigenetic inheritance may only affect fitness if it predictably alters a trait under selection. Evidence has been forwarded that environmental stimuli are important agents in the alteration of epigenes. Ironically, Darwinian evolution may act on these neo-Lamarckian acquired characteristics as well as the cellular mechanisms producing them (e.g. methyltransferase genes). Epigenetic inheritance may confer a fitness benefit to organisms that deal with environmental changes at intermediate timescales.[56] Short-cycling changes are likely to have DNA-encoded regulatory processes, as the probability of the offspring needing to respond to changes multiple times during their lifespans is high. On the other end, natural selection will act on populations experiencing changes on longer-cycling environmental changes. In these cases, if epigenetic priming of the next generation is deleterious to fitness over most of the interval (e.g. misinformation about the environment), these genotypes and epigenotypes will be lost. For intermediate time cycles, the probability of the offspring encountering a similar environment is sufficiently high without substantial selective pressure on individuals lacking a genetic architecture capable of responding to the environment. Naturally, the absolute lengths of short, intermediate, and long environmental cycles will depend on the trait, the length of epigenetic memory, and the generation time of the organism. Much of the interpretation of epigenetic fitness effects centers on the hypothesis that epigenes are important contributors to phenotypes, which remains to be resolved.
Deleterious effects
Inherited epigenetic marks may be important for regulating important components of fitness. In plants, for instance, the Lcyc gene in Linaria vulgaris controls the symmetry of the flower. Linnaeus first described radially symmetric mutants, which arise when Lcyc is heavily methylated.[57] Given the importance of floral shape to pollinators,[58] methylation of Lcyc homologues (e.g. CYCLOIDEA) may have deleterious effects on plant fitness. In animals, numerous studies have shown that inherited epigenetic marks can increase susceptibility to disease. Transgenerational epigenetic influences are also suggested to contribute to disease, especially cancer, in humans. Tumor methylation patterns in gene promoters have been shown to correlate positively with familial history of cancer.[59] Furthermore, methylation of the MSH2 gene is correlated with early-onset colorectal and endometrial cancers.[60]
Putatively adaptive effects
Experimentally demethylated seeds of the model organism Arabidopsis thaliana have significantly higher mortality, stunted growth, delayed flowering, and lower fruit set,[61] indicating that epigenes may increase fitness. Furthermore, environmentally induced epigenetic responses to stress have been shown to be inherited and positively correlated with fitness.[62] In animals, communal nesting changes mouse behavior increasing parental care regimes[63] and social abilities[64] that are hypothesized to increase offspring survival and access to resources (such as food and mates), respectively.
Macroevolutionary patterns
Inherited epigenetic effects on phenotypes have been documented in bacteria, protists, fungi, plants, and animals.[27] Though no systematic study of epigenetic inheritance has been conducted (most focus on model organisms), there is preliminary evidence that this mode of inheritance is more important in plants than in animals.[27] The early differentiation of animal germlines is likely to preclude epigenetic marking occurring later in development, while in plants and fungi somatic cells may be incorporated into the germ line.[65][66]
Life history patterns may also contribute to the occurrence of epigenetic inheritance. Sessile organisms, those with low dispersal capability, and those with simple behavior may benefit most from conveying information to their offspring via epigenetic pathways. Geographic patterns may also emerge, where highly variable and highly conserved environments might host fewer species with important epigenetic inheritance.
Controversies
Humans have long recognized that traits of the parents are often seen in offspring. This insight led to the practical application of selective breeding of plants and animals, but did not address the central question of inheritance: how are these traits conserved between generations, and what causes variation? Several positions have been held in the history of evolutionary thought.
Blending vs. particulate inheritance
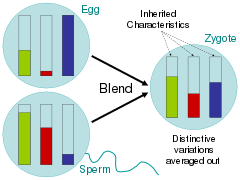
Addressing these related questions, scientists during the time of the Enlightenment largely argued for the blending hypothesis, in which parental traits were homogenized in the offspring much like buckets of different colored paint being mixed together.[67] Critics of Charles Darwin's On the Origin of Species, pointed out that under this scheme of inheritance, variation would quickly be swamped by the majority phenotype.[68] In the paint bucket analogy, this would be seen by mixing two colors together and then mixing the resulting color with only one of the parent colors 20 times; the rare variant color would quickly fade.
Unknown to most of the European scientific community, a monk by the name of Gregor Mendel had resolved the question of how traits are conserved between generations through breeding experiments with pea plants.[69] Charles Darwin thus did not know of Mendel's proposed "particulate inheritance" in which traits were not blended but passed to offspring in discrete units that we now call genes. Darwin came to reject the blending hypothesis even though his ideas and Mendel's were not unified until the 1930s, a period referred to as the modern synthesis.
Inheritance of innate vs. acquired characteristics
In his 1809 book, Philosophie Zoologique,[70] Jean-Baptiste Lamarck recognized that each species experiences a unique set of challenges due to its form and environment. Thus, he proposed that the characters used most often would accumulate a "nervous fluid." Such acquired accumulations would then be transmitted to the individual's offspring. In modern terms, a nervous fluid transmitted to offspring would be a form of epigenetic inheritance.
Lamarckism, as this body of thought became known, was the standard explanation for change in species over time when Charles Darwin and Alfred Russel Wallace co-proposed a theory of evolution by natural selection in 1859. Responding to Darwin and Wallace's theory, a revised neo-Lamarckism attracted a small following of biologists,[71] though the Lamarckian zeal was quenched in large part due to Weismann's[72] famous experiment in which he cut off the tails of mice over several successive generations without having any effect on tail length. Thus the emergent consensus that acquired characteristics could not be inherited became canon.[2]
Revision of evolutionary theory
Non-genetic variation and inheritance, however, proved to be quite common. Concurrent with the 20th-century development of the modern evolutionary synthesis (unifying Mendelian genetics and natural selection), C. H. Waddington (1905-1975) was working to unify developmental biology and genetics. In so doing, he adopted the word "epigenetic"[73] to represent the ordered differentiation of embryonic cells into functionally distinct cell types despite having identical primary structure of their DNA.[74] Researchers discussed Waddington's epigenetics sporadically - it became more of a catch-all for puzzling non-genetic heritable characters rather than a concept advancing the body of inquiry.[75][76] Consequently, the definition of Waddington's word has itself evolved, broadening beyond the subset of developmentally signaled, inherited cell specialization.
Some scientists have questioned whether epigenetic inheritance compromises the foundation of the modern synthesis. Outlining the central dogma of molecular biology, Francis Crick[77] succinctly stated, "DNA is held in a configuration by histone[s] so that it can act as a passive template for the simultaneous synthesis of RNA and protein[s]. None of the detailed 'information' is in the histone." However, he closes the article stating, "this scheme explains the majority of the present experimental results!" Indeed, the emergence of epigenetic inheritance (in addition to advances in the study of evolutionary-development, phenotypic plasticity, evolvability, and systems biology) has strained the current framework of the modern evolutionary synthesis, and prompted the re-examination of previously dismissed evolutionary mechanisms.[78]
There has been much critical discussion of mainstream evolutionary theory by Edward J Steele, Robyn A Lindley and colleagues,[79][80][81][82][83] Fred Hoyle and N. Chandra Wickramasinghe,[84][85][86] Yongsheng Liu[87][88] Denis Noble,[89][90] John Mattick[91] and others that the logical inconsistencies as well as Lamarckian Inheritance effects involving direct DNA modifications, as well as the just described indirect, viz. epigenetic, transmissions, challenge conventional thinking in evolutionary biology and adjacent fields.
See also
References
- Bradbury J (December 2003). "Human epigenome project--up and running". PLOS Biology. 1 (3): E82. doi:10.1371/journal.pbio.0000082. PMC 300691. PMID 14691553.
- Moore DS (2015). The Developing Genome. Oxford University Press. ISBN 978-0-19-992234-5.
- Heard E, Martienssen RA (2014). "Transgenerational epigenetic inheritance: myths and mechanisms". Cell. 157 (1): 95–109. doi:10.1016/j.cell.2014.02.045. PMC 4020004. PMID 24679529.CS1 maint: uses authors parameter (link)
- Jablonka E and MJ Lamb (2010). Transgenerational epigenetic inheritance. In: M Pigliucci and GB Müller Evolution, the expanded synthesis
- Zordan RE, Galgoczy DJ, Johnson AD (August 2006). "Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop". Proceedings of the National Academy of Sciences of the United States of America. 103 (34): 12807–12. doi:10.1073/pnas.0605138103. PMC 1535343. PMID 16899543.
- Beisson J, Sonneborn TM (February 1965). "Cytoplasmic Inheritance of the Organization of the Cell Cortex in Paramecium Aurelia". Proceedings of the National Academy of Sciences of the United States of America. 53 (2): 275–82. Bibcode:1965PNAS...53..275B. doi:10.1073/pnas.53.2.275. PMC 219507. PMID 14294056.
- Soto C, Castilla J (July 2004). "The controversial protein-only hypothesis of prion propagation". Nature Medicine. 10 Suppl (7): S63–7. doi:10.1038/nm1069. PMID 15272271.
- Vastenhouw NL, Brunschwig K, Okihara KL, Müller F, Tijsterman M, Plasterk RH (August 2006). "Gene expression: long-term gene silencing by RNAi". Nature. 442 (7105): 882. Bibcode:2006Natur.442..882V. doi:10.1038/442882a. PMID 16929289.
- Bond DM, Finnegan EJ (2007). "Passing the message on: inheritance of epigenetic traits". Trends in Plant Science. 12 (5): 211–6. doi:10.1016/j.tplants.2007.03.010. PMID 17434332.
- Morison IM, Reeve AE (1998). "A catalogue of imprinted genes and parent-of-origin effects in humans and animals". Human Molecular Genetics. 7 (10): 1599–609. doi:10.1093/hmg/7.10.1599. PMID 9735381.
- Scott RJ, Spielman M, Bailey J, Dickinson HG (September 1998). "Parent-of-origin effects on seed development in Arabidopsis thaliana". Development. 125 (17): 3329–41. PMID 9693137.
- Adenot PG, Mercier Y, Renard JP, Thompson EM (1997). "Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos" (PDF). Development. 124 (22): 4615–25. PMID 9409678.
- Santos F, Hendrich B, Reik W, Dean W (2002). "Dynamic reprogramming of DNA methylation in the early mouse embryo". Developmental Biology. 241 (1): 172–82. doi:10.1006/dbio.2001.0501. PMID 11784103.
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J (April 2000). "Active demethylation of the paternal genome in the mouse zygote". Current Biology. 10 (8): 475–8. doi:10.1016/S0960-9822(00)00448-6. PMID 10801417.
- Fulka H, Mrazek M, Tepla O, Fulka J (2004). "DNA methylation pattern in human zygotes and developing embryos". Reproduction (Cambridge, England). 128 (6): 703–8. doi:10.1530/rep.1.00217. PMID 15579587.
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA (January 2013). "Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine". Science. 339 (6118): 448–52. Bibcode:2013Sci...339..448H. doi:10.1126/science.1229277. PMC 3847602. PMID 23223451.
- Surani MA, Hajkova P (2010). "Epigenetic reprogramming of mouse germ cells toward totipotency". Cold Spring Harbor Symposia on Quantitative Biology. 75: 211–8. doi:10.1101/sqb.2010.75.010. PMID 21139069.
- Zhang Z, Shibahara K, Stillman B (November 2000). "PCNA connects DNA replication to epigenetic inheritance in yeast". Nature. 408 (6809): 221–5. Bibcode:2000Natur.408..221Z. doi:10.1038/35041601. PMID 11089978.
- Henderson DS, Banga SS, Grigliatti TA, Boyd JB (1994). "Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophila gene encoding PCNA". The EMBO Journal. 13 (6): 1450–9. doi:10.1002/j.1460-2075.1994.tb06399.x. PMC 394963. PMID 7907981.
- Probst AV, Dunleavy E, Almouzni G (2009). "Epigenetic inheritance during the cell cycle". Nature Reviews Molecular Cell Biology. 10 (3): 192–206. doi:10.1038/nrm2640. PMID 19234478.
- Morgan HD, Santos F, Green K, Dean W, Reik W (2005). "Epigenetic reprogramming in mammals". Human Molecular Genetics. 14 (Review Issue 1): R47–58. doi:10.1093/hmg/ddi114. PMID 15809273.
- Santos F, Peters AH, Otte AP, Reik W, Dean W (2005). "Dynamic chromatin modifications characterise the first cell cycle in mouse embryos". Developmental Biology. 280 (1): 225–36. doi:10.1016/j.ydbio.2005.01.025. PMID 15766761.
- Taguchi YH (2015). "Identification of aberrant gene expression associated with aberrant promoter methylation in primordial germ cells between E13 and E16 rat F3 generation vinclozolin lineage". BMC Bioinformatics. 16 Suppl 18: S16. doi:10.1186/1471-2105-16-S18-S16. PMC 4682393. PMID 26677731.
- Richards EJ (2006). "Inherited epigenetic variation--revisiting soft inheritance". Nature Reviews Genetics. 7 (5): 395–401. doi:10.1038/nrg1834. PMID 16534512.
- Jablonka E, Lamb MJ (1998). "Epigenetic inheritance in evolution". Journal of Evolutionary Biology. 11 (2): 159–183. doi:10.1046/j.1420-9101.1998.11020159.x.
- Bird A, Kirschner M, Gerhart J, Moore T, Wopert L (1998). "Comments on "Epigenetic inheritance in evolution"". Journal of Evolutionary Biology. 11 (2): 185–188, 213–217, 229–232, 239–240. doi:10.1046/j.1420-9101.1998.11020185.x.
- Jablonka E, Raz G (2009). "Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution". The Quarterly Review of Biology. 84 (2): 131–76. CiteSeerX 10.1.1.617.6333. doi:10.1086/598822. PMID 19606595.
- Rassoulzadegan M, Cuzin F (2015). "Epigenetic heredity: RNA-mediated modes of phenotypic variation". Ann N Y Acad Sci. 1341 (1): 172–5. Bibcode:2015NYASA1341..172R. doi:10.1111/nyas.12694. PMID 25726734.
- Bossdorf O, Richards CL, Pigliucci M (2008). "Epigenetics for ecologists". Ecology Letters. 11 (2): 106–115. doi:10.1111/j.1461-0248.2007.01130.x. PMID 18021243.
- Molinier J, Ries G, Zipfel C, Hohn B (August 2006). "Transgeneration memory of stress in plants". Nature. 442 (7106): 1046–9. Bibcode:2006Natur.442.1046M. doi:10.1038/nature05022. PMID 16892047.
- Coe, E. H. (June 1959). "A Regular and Continuing Conversion-Type Phenomenon at the B Locus in Maize". Proceedings of the National Academy of Sciences of the United States of America. 45 (6): 828–832. Bibcode:1959PNAS...45..828C. doi:10.1073/pnas.45.6.828. ISSN 0027-8424. PMC 222644. PMID 16590451.
- Chandler, Vicki L. (2007-02-23). "Paramutation: From Maize to Mice". Cell. 128 (4): 641–645. doi:10.1016/j.cell.2007.02.007. ISSN 1097-4172. PMID 17320501.
- Stam, Maike; Belele, Christiane; Ramakrishna, Wusirika; Dorweiler, Jane E; Bennetzen, Jeffrey L; Chandler, Vicki L (October 2002). "The regulatory regions required for B' paramutation and expression are located far upstream of the maize b1 transcribed sequences". Genetics. 162 (2): 917–930. ISSN 0016-6731. PMC 1462281. PMID 12399399.
- Chandler, Vicki L.; Arteaga-Vazquez, Mario A.; Bader, Rechien; Stam, Maike; Sidorenko, Lyudmila; Belele, Christiane L. (2013-10-17). "Specific Tandem Repeats Are Sufficient for Paramutation-Induced Trans-Generational Silencing". PLOS Genetics. 9 (10): e1003773. doi:10.1371/journal.pgen.1003773. ISSN 1553-7404. PMC 3798267. PMID 24146624.
- Arteaga-Vazquez, Mario; Sidorenko, Lyudmila; Rabanal, Fernando A.; Shrivistava, Roli; Nobuta, Kan; Green, Pamela J.; Meyers, Blake C.; Chandler, Vicki L. (2010-07-20). "RNA-mediated trans-communication can establish paramutation at the b1 locus in maize". Proceedings of the National Academy of Sciences of the United States of America. 107 (29): 12986–12991. Bibcode:2010PNAS..10712986A. doi:10.1073/pnas.1007972107. ISSN 0027-8424. PMC 2919911. PMID 20616013.
- Stam, Maike; Laat, Wouter de; Driel, Roel van; Haring, Max; Bader, Rechien; Louwers, Marieke (2009-03-01). "Tissue- and Expression Level–Specific Chromatin Looping at Maize b1 Epialleles". The Plant Cell. 21 (3): 832–842. doi:10.1105/tpc.108.064329. ISSN 1532-298X. PMC 2671708. PMID 19336692.
- Haring, Max; Bader, Rechien; Louwers, Marieke; Schwabe, Anne; Driel, Roel van; Stam, Maike (2010-08-01). "The role of DNA methylation, nucleosome occupancy and histone modifications in paramutation". The Plant Journal. 63 (3): 366–378. doi:10.1111/j.1365-313X.2010.04245.x. ISSN 1365-313X. PMID 20444233.
- Chandler, Vicki L.; Kermicle, Jerry L.; Hollick, Jay B.; Kubo, Kenneth M.; Carey, Charles C.; Dorweiler, Jane E. (2000-11-01). "mediator of paramutation1 Is Required for Establishment and Maintenance of Paramutation at Multiple Maize Loci". The Plant Cell. 12 (11): 2101–2118. doi:10.1105/tpc.12.11.2101. ISSN 1532-298X. PMC 150161. PMID 11090212.
- Alleman, Mary; Chandler, Vicki (2008-04-01). "Paramutation: Epigenetic Instructions Passed Across Generations". Genetics. 178 (4): 1839–1844. ISSN 1943-2631. PMC 2323780. PMID 18430919.
- Meyers, Blake C.; Chandler, Vicki L.; Green, Pamela J.; Yen, Yang; Jeong, Dong-Hoon; Sidorenko, Lyudmila; Arteaga-Vazquez, Mario; Accerbi, Monica; Paoli, Emanuele De (2008-09-30). "Distinct size distribution of endogenous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant". Proceedings of the National Academy of Sciences. 105 (39): 14958–14963. Bibcode:2008PNAS..10514958N. doi:10.1073/pnas.0808066105. ISSN 1091-6490. PMC 2567475. PMID 18815367.
- Chandler, Vicki L.; Sikkink, Kristin; White, Joshua; Dorweiler, Jane E.; Seshadri, Vishwas; McGinnis, Karen; Sidorenko, Lyudmila; Alleman, Mary (July 2006). "An RNA-dependent RNA polymerase is required for paramutation in maize". Nature. 442 (7100): 295–298. Bibcode:2006Natur.442..295A. doi:10.1038/nature04884. ISSN 1476-4687. PMID 16855589.
- Arteaga-Vazquez, Mario Alberto; Chandler, Vicki Lynn (April 2010). "Paramutation in maize: RNA mediated trans-generational gene silencing". Current Opinion in Genetics & Development. 20 (2): 156–163. doi:10.1016/j.gde.2010.01.008. ISSN 0959-437X. PMC 2859986. PMID 20153628.
- Huang, J.; Lynn, J.S.; Schulte, L.; Vendramin, S.; McGinnis, K. (2017-01-01). Epigenetic Control of Gene Expression in Maize. International Review of Cell and Molecular Biology. 328. pp. 25–48. doi:10.1016/bs.ircmb.2016.08.002. ISBN 9780128122204. ISSN 1937-6448. PMID 28069135.
- Chandler, Vicki L. (2010-10-29). "Paramutation's Properties and Puzzles". Science. 330 (6004): 628–629. Bibcode:2010Sci...330..628C. doi:10.1126/science.1191044. ISSN 1095-9203. PMID 21030647.
- Luo, Lijun; Li, Tiemei; Li, Mingshou; Lou, Qiaojun; Wei, Haibin; Xia, Hui; Chen, Liang; Zheng, Xiaoguo (2017-01-04). "Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant's adaptation to drought condition". Scientific Reports. 7: 39843. Bibcode:2017NatSR...739843Z. doi:10.1038/srep39843. ISSN 2045-2322. PMC 5209664. PMID 28051176.
- Jander, Georg; Felton, Gary W.; Agrawal, Anurag A.; Sun, Joel Y.; Halitschke, Rayko; Tian, Donglan; Casteel, Clare L.; Vos, Martin De; Rasmann, Sergio (2012-02-01). "Herbivory in the Previous Generation Primes Plants for Enhanced Insect Resistance". Plant Physiology. 158 (2): 854–863. doi:10.1104/pp.111.187831. ISSN 1532-2548. PMC 3271773. PMID 22209873.
- Quadrana, Leandro; Colot, Vincent (2016). "Plant Transgenerational Epigenetics". Annual Review of Genetics. 50 (1): 467–491. doi:10.1146/annurev-genet-120215-035254. PMID 27732791.
- Wei Y, Schatten H, Sun QY (2014). "Environmental epigenetic inheritance through gametes and implications for human reproduction". Human Reproduction Update. 21 (2): 194–208. doi:10.1093/humupd/dmu061. PMID 25416302.
- Carl Zimmer (31 Jan 2018). "The Famine Ended 70 Years Ago, but Dutch Genes Still Bear Scars". The New York Times.
- Kate Baggaley (9 Nov 2018). "Civil War study shows father's stress affects son's lifespan".
- Lalande M (1996). "Parental imprinting and human disease". Annual Review of Genetics. 30: 173–95. doi:10.1146/annurev.genet.30.1.173. PMID 8982453.
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ (August 2004). "Epigenetic programming by maternal behavior". Nature Neuroscience. 7 (8): 847–54. doi:10.1038/nn1276. PMID 15220929.
- McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ (March 2009). "Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse". Nature Neuroscience. 12 (3): 342–8. doi:10.1038/nn.2270. PMC 2944040. PMID 19234457.
- Meaney MJ, Szyf M (2005). "Environmental programming of stress responses through DNA methylation: life at the interface between a dynamic environment and a fixed genome". Dialogues in Clinical Neuroscience. 7 (2): 103–23. PMC 3181727. PMID 16262207.
- Radtke KM, Ruf M, Gunter HM, Dohrmann K, Schauer M, Meyer A, Elbert T (July 2011). "Transgenerational impact of intimate partner violence on methylation in the promoter of the glucocorticoid receptor". Translational Psychiatry. 1 (July 19): e21. doi:10.1038/tp.2011.21. PMC 3309516. PMID 22832523.
- Jablonka E, Lamb MJ (2005). Epigenetic inheritance and evolution: the Lamarckian dimension (Reprinted ed.). Oxford: Oxford University Press. ISBN 978-0-19-854063-2.
- Cubas P, Vincent C, Coen E (1999). "An epigenetic mutation responsible for natural variation in floral symmetry". Nature. 401 (6749): 157–61. Bibcode:1999Natur.401..157C. doi:10.1038/43657. PMID 10490023.
- Dafni A, Kevan PG (1997). "Flower size and shape: implications in pollination". Israeli Journal of Plant Science. 45 (2–3): 201–211. doi:10.1080/07929978.1997.10676684.
- Frazier ML, Xi L, Zong J, Viscofsky N, Rashid A, Wu EF, Lynch PM, Amos CI, Issa JP (August 2003). "Association of the CpG island methylator phenotype with family history of cancer in patients with colorectal cancer". Cancer Research. 63 (16): 4805–8. PMID 12941799.
- Chan TL, Yuen ST, Kong CK, Chan YW, Chan AS, Ng WF, Tsui WY, Lo MW, Tam WY, Li VS, Leung SY (October 2006). "Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer". Nature Genetics. 38 (10): 1178–83. doi:10.1038/ng1866. PMID 16951683.
- Bossdorf O, Arcuri D, Richards CL, Pigliucci M (2010). "Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in Arabidopsis thaliana" (PDF). Evolutionary Ecology. 24 (3): 541–553. doi:10.1007/s10682-010-9372-7.
- Whittle CA, Otto SP, Johnston MO, Krochko JE (2009). "Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana". Botany. 87 (6): 650–657. doi:10.1139/b09-030.
- Curley, JP, FA Champagne, and P Bateson (2007) Communal nesting induces alternative emotional, social and maternal behavior in offspring. Society for Behavioral Neuroendocrinology 11th Annual Meeting Pacific Grove, CA, USA. Cited in Branchi I (April 2009). "The mouse communal nest: investigating the epigenetic influences of the early social environment on brain and behavior development". Neuroscience and Biobehavioral Reviews. 33 (4): 551–9. doi:10.1016/j.neubiorev.2008.03.011. PMID 18471879.
- Branchi I, D'Andrea I, Fiore M, Di Fausto V, Aloe L, Alleva E (October 2006). "Early social enrichment shapes social behavior and nerve growth factor and brain-derived neurotrophic factor levels in the adult mouse brain". Biological Psychiatry. 60 (7): 690–6. doi:10.1016/j.biopsych.2006.01.005. PMID 16533499.
- Whitham TG, Slobodchikoff CN (1981). "Evolution by individuals, plant-herbivore interactions, and mosaics of genetic variability: The adaptive significance of somatic mutations in plants". Oecologia. 49 (3): 287–292. Bibcode:1981Oecol..49..287W. doi:10.1007/BF00347587. PMID 28309985.
- Turian G (1979). "Sporogenesis in fungi". Annual Review of Phytopathology. 12: 129–137. doi:10.1146/annurev.py.12.090174.001021.
- Vorzimmer P (1963). "Charles Darwin and blending inheritance". Isis. 54 (3): 371–390. doi:10.1086/349734.
- Jenkin F (1867). "Review of The Origin of Species". North British Review.
- Mendel G (1866). "Versuche über Plflanzenhybriden. Verhandlungen des naturforschenden Vereines in Brünn" [Experiments in Plant Hybridization] (PDF). Read at the February 8th, and March 8th, 1865, meetings of the Brünn Natural History Society (in German).
- Lamarck, JB (1809). Philosophie zoologique: ou Exposition des considérations relative à l'histoire naturelle des animaux. Dentu et L'Auteur, Paris.
- Bowler PJ (1989). Evolution, the history of an idea. Berkeley: University of California Press. ISBN 978-0-520-06386-0.
- Weismann A (1891). Poulton EB, Schönland S, Shipley E (eds.). Essays upon heredity and kindred biological problems. Oxford: Clarendon Press. doi:10.5962/bhl.title.28066.
- {{oed | epigenetic
- Waddington CH (2016) [1939]. "Development as an Epigenetic Process". Introduction to Modern Genetics. London: Allen and Unwin. ISBN 9781317352037.
One of the classical controversies in embryology was that between the preformationists and the epigenisists[sic]. [...] the interaction of these constituents gives rise to new types of tissue and organ which were not present originally, and in so far development must be considered as 'epigenetic.'
- Holliday R (2006). "Epigenetics: a historical overview". Epigenetics. 1 (2): 76–80. doi:10.4161/epi.1.2.2762. PMID 17998809.
- Nanney DL (July 1958). "Epigenetic Control Systems". Proceedings of the National Academy of Sciences of the United States of America. 44 (7): 712–7. Bibcode:1958PNAS...44..712N. doi:10.1073/pnas.44.7.712. PMC 528649. PMID 16590265.
- Crick FH (1958). "On protein synthesis" (PDF). Symposia of the Society for Experimental Biology. 12: 138–63. PMID 13580867.
- Pigliucci M (December 2007). "Do we need an extended evolutionary synthesis?". Evolution; International Journal of Organic Evolution. 61 (12): 2743–9. doi:10.1111/j.1558-5646.2007.00246.x. PMID 17924956.
- Steele, E.J. (1979). Somatic selection and adaptive evolution: on the inheritance of acquired characters (1st edit ed.). Toronto: Williams-Wallace.
- Steele, E.J.; Lindley, R.A.; Blanden, R.V. (1998). Davies, Paul (ed.). Lamarck's signature: how retrogenes are changing Darwin's natural selection paradigm. Frontiers of Science. Sydney: Allen & Unwin.
- Lindley, R.A. (2010). The Soma: how our genes really work and how that changes everything!. Piara Waters, CYO Foundation. ISBN 978-1451525649.
- Steele, E.J.; Lloyd, S.S. (2015). "Soma-to-germline feedback is implied by the extreme polymorphism at IGHV relative to MHC". BioEssays. 37 (5): 557–569. doi:10.1002/bies.201400213. PMID 25810320.
- Steele, E.J. (2016). Levin, Michael; Adams, Dany Spencer (eds.). Origin of congenital defects: stable inheritance through the male line via maternal antibodies specific for eye lens antigens inducing autoimmune eye defects in developing rabbits in utero. Ahead of the Curve -Hidden breakthroughs in the biosciences. Bristol, UK: IOP Publishing Ltd. pp. Chapter 3.
- Hoyle, F.; Wickramasinghe, Chandra (1982). Why neo-Darwinism does not work. Cardiff: University College Cardiff Press. ISBN 0-906449-50-2.
- Hoyle, F.; Wickramasinghe, N.C. (1979). Diseases from space. London: J.M. Dent.
- Hoyle, F.; Wickramasinghe, N.C. (1981). Evolution from space. London: J.M. Dent.
- Liu, Y. (2007). "Like father like son. A fresh review of the inheritance of acquired characteristics". EMBO Reports. 8 (9): 798–803. doi:10.1038/sj.embor.7401060. PMC 1973965. PMID 17767188.
- Liu, Y.; Li, X. (2016). "Darwin's Pangenesis as a molecular theory of inherited diseases". Gene 2016a. 582 (1): 19–22. doi:10.1016/j.gene.2016.01.051. PMID 26836487.
- Noble, D. (2011). "A theory of biological relativity: no privileged level of causation". Interface Focus. 2 (1): 55–64. doi:10.1098/rsfs.2011.0067. PMC 3262309. PMID 23386960.
- Noble, D. (2013). "Physiology is rocking the foundations of evolutionary biology". Exp. Physiol. 98 (8): 1235–1243. doi:10.1113/expphysiol.2012.071134. PMID 23585325.
- Mattick, J.S. (2012). "Rocking the foundations of molecular genetics". Proc Natl Acad Sci USA. 109 (41): 16400–16401. Bibcode:2012PNAS..10916400M. doi:10.1073/pnas.1214129109. PMC 3478605. PMID 23019584.