Torovirus
Torovirus is a genus of viruses in the order Nidovirales, in the family Tobaniviridae, in the subfamily Torovirinae.[1] They primarily infect vertebrates.,[2][3] especially cattle, pig, and horse.[4] Diseases associated with this genus include: gastroenteritis,[4] which commonly presents in mammals,[5] Torovirus is the sole genus in the monotypic subfamily Torovirinae.[6] Torovirus is also a monotypic taxon, containing only one subgenus, Renitovirus.[6]
| Renitovirus | |
|---|---|
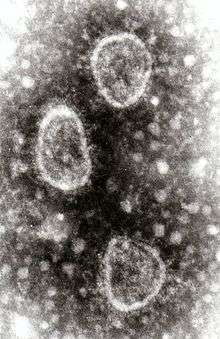 | |
| Torovirus particles | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Family: | Tobaniviridae |
| Subfamily: | Torovirinae |
| Genus: | Torovirus |
| Subgenus: | Renitovirus |
| Type species | |
| Equine torovirus | |
| Species | |
| |
The discovery of the first torovirus can be traced back to 1970s. Equine torovirus (EToV) was accidentally found in the rectal sample from a horse who was suffering from severe diarrhea. The ‘Breda’ bovine torovirus was later found in 1979 while investigation in a dairy farm in Breda. They had several calves suffering from severe diarrhea for months. In 1984, torovirus like particles were detected with Electron Microscope (EM) technique in the human patients with gastroenteritis.[7] Although, the morbidity and the complications due to the torovirus are unwitnessed in humans so far, there is a lot more research required to better understand the behavior of the toroviruses.
History
In 1972, a virus was isolated from a horse in Berne, Switzerland. The virus did not react with antisera against known equine viruses and was shown to have a unique morphology and substructure.[8] In 1982 a similar, unclassified virus was isolated from calves in Breda, Iowa.[9] In 1984 particles resembling these viruses were discovered in the faeces of humans.[10] On the basis of the available information on these viruses, the establishment of a new family of viruses—Toroviridae—was proposed at the 6th International Congress on the Taxonomy of Viruses at Sendai, Japan,[11] but the genus is currently assigned to the subfamily Torovirinae in the family Tobaniviridae, order Nidovirales.[2]
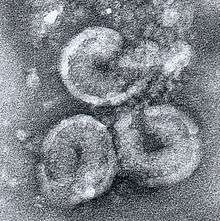
Structure
Torovirus particles share characteristics with members of the related family Coronaviridae; they are round, pleomorphic, enveloped viruses about 120 to 140 nm in diameter. The virus particle has surface spike proteins that are club-shaped and are evenly dispersed over the surface. A nucleocapsid that is doughnut-shaped with helical symmetry is present.[4][12]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Torovirus | Spherical | Helical | Enveloped | Linear | Monopartite |
Torovirus particles typically possess a helical symmetrical nucleocapsid that is coiled into a hollow cylindrical shape. The diameter is approximately 23 nm with an average length of 104 nm, where every turn cycle is at intervals of 4.5 nm.[7]
The different origins of the various torovirus strains has led to the classification of different strains like bovine torovirus (BToV), equine torovirus (EToV) and human torovirus (HuTV) are among most researched. The approximate length of genome of the bovine torovirus is approximately 28.5 kilo-bytes. Based on the genetic characteristics and the morphology of the BToV, researchers derive that the toroviruses possibly share the same ancestry with the coronaviruses.
Life cycle
Viral replication is cytoplasmic. Entry into the host cell is achieved by attachment of the viral S protein (maybe also HE if present) to host receptors, which mediates endocytosis. Replication follows the positive stranded RNA virus replication model. Positive-stranded RNA-virus transcription, using the premature termination model of subgenomic RNA transcription is the method of transcription. Translation takes place by -1 ribosomal frameshifting. Cattle, pig, and horse serve as the natural host. Transmission is thought to be via the faecal-oral route.[4]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Torovirus | Cattle; pig; horse | Epithelium: respiratory; epithelium: enteric | Cell receptor endocytosis | Budding | Cytoplasm | Cytoplasm | Oral-fecal |
Genome
Toroviruses are class IV viruses, and have a single piece of single-stranded, positive-sense RNA. The total length of this is about 28,000 nucleotides and toroviruses have a complex replication mechanism that includes the use of subgenomic mRNA, ribosomal frameshifting, and polymerase stuttering.[13]
The outbreak of Severe Acute Respiratory Syndrome Coronavirus in 2003, created interest in the viruses in the family Coronaviridae. The genus Torovirus which is also said to have the same common ancestor as suggested by their shared characteristics, is less well known at present, because it has not yet been cultivated in the cell culture. Only the Berne Torovirus (BToV) can be cultivated in the cell culture so far. However, researchers have sequenced the complete genome of bovine-Torovirus (BoTV-1). The length of bovine torovirus is 28.475 kb and the genome contains mainly the data for the replicase gene which is approximately 20.2 kb.[14] It contains mainly two open reading frame ORF1a and ORF1b, that encodes for the polyproteins called pp1a and pp1ab.[15] No disease has been confirmed with the BToV in humans so far, however, various antibodies relating to the BToV has been identified in various mammals, which suggest that the extent of spread of torovirus is wide.
Evolution
Porcine Torovirus evolved ~1951.[16] Initially, toroviruses were thought to be a new family with enveloped RNA. However, later studies revealed its morphological and behavioral ties with the coronavirus family i.e. Coronaviridae.[17] More distantly, it can also be genetically linked to the Arteriviruses (Arteriviridae). The research done on the torovirus illustrates the divergence from a common ancestor. There are numerous studies done in regard to the toroviruses. The samples of the virus RNA is usually extracted from the fecal specimens from different species. Sequencing various structural proteins give the researchers the diversion from one to another. Major torovirus strains were found by sequence studies of structural viral protein like S, M, HE, N.[18] Toroviruses like bovine torovirus (BToV) Breda, porcine torovirus (PToV) Markelo, equine torovirus Berne, and the putative human torovirus are classified with the sequencing techniques. The origin of these virus strains can be attributed to the evolution in these structural genes involving various recombination events in genetic recombination and multiple unknown mutations over time. The structural and behavioral characteristics of Toroviruses coincide with other viruses like Arteriviruses and Coronaviruses, hence scientists derive the common ancestry relationship of these viruses.
Taxonomy
Until recently, the toroviruses were not assigned any family. The recent molecular analysis of the virus revealed its similarities with Arterivirus and coronaviruses, which led to the inclusion of the Torovirus along with the Arterivirus in the previously monogeneric Coronaviridae.[19] At present, toroviruses are included in the order Nidovirales sub family Torovirinae, family Tobaniviridae. Resemblance, molecular and genetic similarities, virion dimensions, behavioral ties and other characteristic similarities and differences are observed by the researchers for the taxonomic classification of the virus. In the toroviruses, the Berne virus has been extensively studied at the molecular level as compared to its other members. In 1992, the International Committee on Taxonomy of Viruses ICTV got enough data to consider torovirus in the coronavirus family due to the similarities in structure, replication behavior and the genetic sequencing.
Human torovirus
In 1984, torovirus like particles were observed in human patients suffering from gastroenteritis or severe diarrhea.[20][21] A number of case studies then started to emerge from various parts of the world. Torovirus like particles (ToVL) was reported in scientific studies from USA, France, The Netherlands, Canada, Great Britain, India and Brazil.[22] The mainly presence of the ToVL was reported in children and adults with severe diarrhea. The term Human torovirus (HuTV)is often used to describe the ToVL particles. Because there is much similarity in HuTV and BoTV, there are certain criterion that is followed in the detecting and differentiation of both strains. Numerous studies have been conducted in the past to relate the toroviruses and its pathogenicity. Toroviruses has been found in various intestinal diseases in the children as well as the adults. A study of fecal excretion of Torovirus concluded that out of 206 examined cases, in around 72 (35%) cases the torovirus was found. As compared to the infections by rotavirus or torovirus, Toroviruses were more frequently found in the people that are more immunocompromised. The torovirus infections were characterized by reduced vomiting and increased bloody diarrhea. The immune system’s antibody response mainly developed in the grown-up children who were non-immunocompromised.[23] In addition to gastroenteritis, toroviruses has also been found in the infants with necrotizing enterocolitis (NEC).[24] However, in the same study, the severity of illness and mortality was not much affected in the torovirus positive patients as compared to the torovirus negative NEC patients.
Antigenic Properties and Pathogenicity
Bovine Toroviruses are proposed to have mainly two different serotypes: bovine torovirus serotype 1 (BoTV-1) and bovine torovirus serotype 2 (BoTV-2).[25] Both serotypes of BoTV possess a hemagglutinin that reacts with erythrocytes from mice and rats, but not with human erythrocytes. The BoTV does not elute from rat erythrocytes after 90 min at 36 °C. Both the serotypes of the bovine Torovirus possess a hemagglutinin that reacts with red blood cells in the rodents usually.[26] No evidence so far suggests that reaction with the human erythrocytes. Although, many recent studies revealed the presence of the torovirus in humans associated with many other enteric infections, diarrhea and conditions like gastroenteritis.
The exact mechanism by which the virus induces diarrhea is currently unknown, but the studies reveal that it could be due to infection and death of cells in the small intestine and villi crypts as well as the surface crypt enterocytes in the large intestine. It is also said that the watery diarrhea could be due to lesions in the colon that lead to reduction of water absorption by the cells in the large intestine. The pathogenicity of the torovirus presence has been widely studied and explored in the bovine species, especially calves in the initial stage of life around 4 to 6 months of age.[26] Once the BoTV has been inoculated by an animal orally or nasally, it infects the epithelial cells of the villi and then extends to the components of the digestive system like the large intestine in the crypts of jejunum, ileum and colon. It ultimately results in diarrhea with in 24–72 hours of infection.[27][28] The antigens of BToV has also been reported in the dome epithelial cells and Microfold cells, present in GALT of the peyer’s patches in small intestine.[29] Some researchers suggest that the BToV only infects absorptive enterocytes. However, there are researchers that also suggest that the viral replication could start in the immature epithelial cells of the crypts and can further spread to the villi.[30]
Clinical signs and diagnosis
In cattle, the disease causes diarrhoea and systemic signs such as pyrexia, lethargy and anorexia. In calves, it may cause neurological signs and lead to death in some cases. Many diagnostic techniques for torovirus infection in clinical specimens are now available such as hemagglutination (HA), enzyme-linked immunosorbent assay (ELISA), immune electron microscopy ,hemagglutination-inhibition tests (HA/HI), and nucleic acid hybridization.[31] Mostly torovirus infecting humans are probably closely related to BRV or BEV and relate to any previous history of some enteric disease or infection. It is also the reason that the molecular techniques are considered more promising than the serological assays. Pigs can be infected without showing any signs and symptoms that are physically visible.
Diagnosis of the viral infection involves electron microscopy, ELISA or haemagglutination inhibition.
Treatment and control
Supportive treatment may be given to prevent dehydration and secondary infections. Control relies on good biosecurity measures including prompt isolation and disinfection of premises. As torovirus infections usually interrelates with severe diarrhea, it often led to dehydration. The most common treatment includes giving liquid therapies to the younger patients as the infection is most common in younger human population as well as in cattle. There are no as such specific preventive measures for the Torovirus infection control. Good hygiene and biosecurity practices are effective in prevention of the torovirus infection. Moreover, Antibody-containing colostrum can be given to the patients with estimated dose of 500ml/day.[32] The natural course of infection mostly researched is done in the cattle as the virus origin is related to the enteric disease in the cattle. In an experiment associated with the infection of cattle gave the results that the virus commonly transmit itself under conventional conditions. The initial symptoms in the calves showed the development of diarrhea in first 2 to 3 days of the infection. Most of the calves had the mild dehydration and a few developed a mild fever. However, none of the cases required any therapeutic intervention and were treated usually with the changes in the diet of the infected calves.[33] A research by (Vanopdenbosch et al., 1992a) published a study that torovirus respiratory infections emerges mainly in the first month of life and from 4 to 6 months of age in the autumn season peak. In all those infections, about 25% infections lead to sudden death. Besides other causes like diarrhea, pneumonia and respiratory problems, central nervous system related symptoms have also been reported in some incidental cases.[34] Studies also reveals that the young calves infected with the torovirus need to be given fluid therapies as their body cold face severe dehydration during the infection. However, the adults recover by their immune response without any external treatment if any additional infections are not present. Many researchers also suggest that the disinfection and heat sterilization could easily destroy the virus but no scientific data or reports of such results are available so far.
References
- ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, Perlman S, Poon L, Rottier PJ, Talbot PJ, Woo PC, Ziebuhr J (2011). "Family Coronaviridae". In: Ninth Report of the International Committee on Taxonomy of Viruses. (AMQ King, E Lefkowitz, MJ Adams, and EB Carstens, eds), Elsevier, Oxford, pp. 806-828. ISBN 978-0-12-384684-6.
- de Groot RJ (2007). "Molecular Biology and Evolution of Toroviruses". In: The Nidoviruses. (E.J. Snijder, T. Gallagher and S. Perlman, eds), ASM press, Washington DC, USA, pp. 133-146. ISBN 978-0-387-33012-9.
- "Viral Zone". ExPASy. Retrieved 15 June 2015.
- Desk Encyclopedia of General Virology. Boston: Academic Press. 2009. p. 507. ISBN 978-0-12-375146-1.
- Virus Taxonomy: 2018 Release, International Committee on Taxonomy of Viruses, retrieved 7 December 2018
- Cho, K. O., & Hoet, A. E. (2014). Toroviruses (Coronaviridae). Reference Module in Biomedical Sciences, B978-0-12-801238-3.02674-X. https://doi.org/10.1016/B978-0-12-801238-3.02674-X
- Weiss M, Steck F, Horzinek MC (September 1983). "Purification and partial characterization of a new enveloped RNA virus (Berne virus)" (PDF). J. Gen. Virol. 64 (9): 1849–58. doi:10.1099/0022-1317-64-9-1849. PMID 6886677. Retrieved 11 July 2020.
- Woode GN, Reed DE, Runnels PL, Herrig MA, Hill HT (July 1982). "Studies with an unclassified virus isolated from diarrheic calves". Vet. Microbiol. 7 (3): 221–40. doi:10.1016/0378-1135(82)90036-0. PMC 7117454. PMID 7051518.
- Beards GM, Hall C, Green J, Flewett TH, Lamouliatte F, Du Pasquier P (May 1984). "An enveloped virus in stools of children and adults with gastroenteritis that resembles the Breda virus of calves". Lancet. 1 (8385): 1050–2. doi:10.1016/S0140-6736(84)91454-5. PMC 7173266. PMID 6143978.
- Horzinek MC, Weiss M (October 1984). "Toroviridae: a taxonomic proposal". Zentralblatt für Veterinärmedizin. Reihe B. 31 (9): 649–59. doi:10.1111/j.1439-0450.1984.tb01348.x. hdl:1874/3490. PMID 6393658.
- ICTVdB Management (2006). 03.019.0.02. Torovirus. In: ICTVdB - The Universal Virus Database, version 4. Büchen-Osmond, C. (Ed), Columbia University, New York, USA
- Snijder EJ, Horzinek MC (November 1993). "Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily" (PDF). J. Gen. Virol. 74 (11): 2305–16. doi:10.1099/0022-1317-74-11-2305. PMID 8245847. Retrieved 11 July 2020.
- Ryan Draker, Rachel L. Roper, Martin Petric, Raymond Tellier. The complete sequence of the bovine torovirus genome. Virus Research. Volume 115, Issue 1. 2006. Pages 56-68. ISSN 0168-1702. https://doi.org/10.1016/j.virusres.2005.07.005.
- Ryan Draker, Rachel L. Roper, Martin Petric, Raymond Tellier. The complete sequence of the bovine torovirus genome. Virus Research. Volume 115, Issue 1. 2006. Pages 56-68. ISSN 0168-1702. https://doi.org/10.1016/j.virusres.2005.07.005
- Cong Y, Zarlenga DS, Richt JA, Wang X, Wang Y, Suo S, Wang J, Ren Y, Ren X (2013) Evolution and homologous recombination of the hemagglutinin-esterase gene sequences from porcine Torovirus. Virus Genes
- Snijder. E., Horzinek. Marian. November 1, 1993. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. Journal of General Virology. https://doi.org/10.1099/0022-1317-74-11-2305
- S. L. Smits, A. Lavazza, K. Matiz, M. C. Horzinek, M. P. Koopmans, R. J. de Groot Journal of Virology Aug 2003, 77 (17) 9567-9577; DOI: 10.1128/JVI.77.17.9567-9577.2003
- Cavanagh, D., Brien, D. A., Brinton, M., Enjuanes, L., Holmes, K. V., Horzinek, M. C., Lai, M., Laude, H., Plagemann, P., Siddell, S., Spaan, W., Taguchi, F., & Talbot, P. J. (1994). Revision of the taxonomy of the Coronavirus, Torovirus and Arterivirus genera. Archives of virology, 135(1-2), 227–237. https://doi.org/10.1007/BF01309782
- Beards GM, Green J, Hall C, Flewett TH, Lamouliatte F and du Pasquier P (1984). An enveloped virus in stools of children and adults with gastroenteritis that resembles the Breda virus of calves. Lancet i: 1050–1052.
- Beards GM, Brown DW, Green J, Flewett TH (September 1986). "Preliminary characterisation of torovirus-like particles of humans: comparison with Berne virus of horses and Breda virus of calves". Journal of Medical Virology. 20 (1): 67–78. doi:10.1002/jmv.1890200109. PMC 7166937. PMID 3093635.
- Hoet, A., & Saif, L. (2004). Bovine torovirus (Breda virus) revisited. Animal Health Research Reviews, 5(2), 157-171. doi:10.1079/AHR200498
- Frances B. Jamieson, Elaine E. L. Wang, Cindy Bain, Jennifer Good, Lynn Duckmanton, Martin Petric, Human Torovirus: A New Nosocomial Gastrointestinal Pathogen, The Journal of Infectious Diseases, Volume 178, Issue 5, November 1998, Pages 1263–1269, https://doi.org/10.1086/314434
- Lodha, A., de Silva, N., Petric, M. and Moore, A.M. (2005), Human torovirus: A new virus associated with neonatal necrotizing enterocolitis. Acta Pædiatrica, 94: 1085-1088. doi:10.1111/j.1651-2227.2005.tb02049.x
- Woode GN, Mohammed KA, Saif LJ, Winand NJ, Quesada M, Kelso NE and Pohlenz JF (1983). Diagnostic methods for the newly discovered ‘Breda’ group of calf enteritis inducing viruses. In: Proceedings of the Third International Symposium of Veterinary Laboratory Diagnosticians, Volume 2, pp. 533–538. Ames, USA.
- Woode GN, Reed DE, Runnels PL, Herrig MA and Hill HT (1982). Studies with an unclassified virus isolated from diarrheic calves. Veterinary Microbiology 7: 221–240.
- Pohlenz JFL, Cheville NF, Woode GN and Mokresh AH (1984). Cellular lesions in intestinal mucosa of gnotobiotic calves experimentally infected with a new unclassified bovine virus (Breda virus). Veterinary Pathology 21: 407–417
- Hall GA (1987). Comparative pathology of infection by novel diarrhoea viruses. In: Brock G and Whelan J, editors. Novel Diarrhoea Viruses. Ciba Foundation Symposium, No. 128. Chichester: John Wiley & Sons, pp. 192–217
- Pohlenz JFL, Cheville NF, Woode GN and Mokresh AH (1984). Cellular lesions in intestinal mucosa of gnotobiotic calves experimentally infected with a new unclassified bovine virus (Breda virus). Veterinary Pathology 21: 407–417.
- Koopmans M and Horzinek MC (1995). The pathogenesis of torovirus infections in animals and humans. In: Siddell SG, editor. The Coronaviridae. New York: Plenum Press, pp. 403–413
- Koopmans, M., Herrewegh, A., & Horzinek, M. C. (1991). Diagnosis of torovirus infection. Lancet, 337(8745), 859. https://doi.org/10.1016/0140-6736(91)92573-k
- Woode GN (1990). Breda virus. In: Dinter Z and Morein B, editors. Virus Infections of Ruminants. 3rd edn. Sweden: Elsevier Science, pp. 311–316
- Bosch, A., Rosa M. Pintó, and Abad, Xavier. June, 2013.Survival and Transport of Enteric Viruses in Environment. http://www.ub.edu/virusenterics/wp-content/uploads/2013/06/GOY6.pdf
- Vanopdenbosch, E., Wellemans, G., Oudewater, J., nd Petroff, k. (1992a). Vlaams Di-ergennesk. Tijdschr. 61, 1-7.
External links
- Toroviruses reviewed and published by Wikivet, accessed 08/10/2011
- Viralzone: Torovirus
- Virus Pathogen Database and Analysis Resource (ViPR): Coronaviridae
- ICTV