C2orf73
Uncharacterized protein C2orf73 is a protein that in humans is encoded by the C2orf73 gene. The protein is predicted to be localized to the nucleus.
| C2orf73 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | C2orf73, chromosome 2 open reading frame 73 | ||||||||||||||||||||||||
| External IDs | MGI: 1922337 HomoloGene: 18988 GeneCards: C2orf73 | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) | |||||||||||||||||||||||||
| Location (UCSC) | Chr 2: 54.33 – 54.38 Mb | Chr 11: 30.43 – 30.47 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Gene
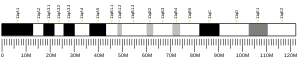
The full gene spans a total of 53,712 base pairs and contains nine exons. The gene's location in the Human genome is on chromosome 2 at position 2p16.2 and is flanked by the genes ACYP2 and SPTBN1.[5] There are no aliases for this gene.
mRNA
The primary mRNA produced by the C2or73 gene is 1921 nucleotides long. There are six other mRNA isoforms produced by alternative splicing and variation in exon length.[6]
| Isoform | Exons | mRNA Length (bases) |
|---|---|---|
| Primary | 2, 3, 5, 6, 7 | 1921 |
| X1 | 2, 3, 5, 6, 7 (truncated), 8, 9 | 1726 |
| X2 | 2, 3 (truncated), 5, 6, 7 (truncated) | 971 |
| X3 | 2 (truncated), 5, 6, 7 (truncated) | 868 |
| X4 | 4, 5, 6, 7 (truncated) | 951 |
| X5 | 1, 5, 6, 7 (truncated) | 1049 |
| X6 | 2, 3, 5, 7 (truncated) | 1034 |
Protein
The protein has a molecular mass of 32,142 Daltons.[7] There are four protein isoforms. The primary isoform (X1) is 287 amino acids long.[8]
C2orf73 contains a short sequence motif, GDWWSH (This motif does not yet have any known function). The protein is lysine rich and leucine poor compared to the content of the average Human gene and has a predicted isoelectric point of 9.305.[9]
| Isoform | From mRNA Isoform | Length (Amino Acids) | Molecular Weight (kDa) | Isoelectric Point |
|---|---|---|---|---|
| X1 | Primary, X1 | 287 | 32.1 | 9.305 |
| X2 | X2 | 229 | 25.4 | 9.120 |
| X3 | X3, X4, X5 | 166 | 18.1 | 9.703 |
| X4 | X6 | 143 | 16.7 | 8.790 |
Structure
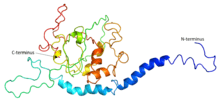
A 3D structure for C2orf73 has not yet been determined experimentally. A computational prediction made by I-TASSER is presented to the right.[10]
The PELE tool on Biology Workbench predicts three likely α-helices and one β-strand in the protein.[14]
Post translational modifications
The GPS, NetPhos, MyHits and SUMOsp tools on ExPASy[15] predict potential post-translational modifications for the protein. Six potential phosphorylation sites and one sumoylation site are predicted.
Subcellular localization
PSORT II predicts C2orf73 to be localized to the nucleus.[16] This is supported by the predicted presence of a sumoylation site, which is involved in nuclear cytoplasmic transport.[17]
Expression
GEO profiles from NCBI show that C2orf73 is weakly expressed in the following tissues in Humans: bone marrow, liver, heart, lung, brain, spinal cord, skeletal muscle, thymus, and epithelium.[18]
Regulation of expression
The Genomatix El Dorado tool predicts many transcription factors to have a high binding affinity in the 1100 base pairs upstream of C2orf73. Many of the transcription factors normally regulate processes such as cell development and differentiation, cell death, and the cell cycle.[19]
Interacting Proteins
Three proteins have been experimentally determined to interact with C2orf73 through Yeast Two-Hybrid experiments.[20]
- FCH and Double SH3 Domains 2 (FCHSD2) - Function has not yet been defined
- Heat Shock Protein Family B Member 1 (HSPB1) - Aids cell's resistance to stress
- SH3 Domain Binding Protein 4 (SH3BP4) - Involved in endocytosis of specific cell surface receptors
Function
The function of C2orf73 is currently not well understood by the scientific community or anyone else.
Homology
There are no paralogs of C2orf73 in the Human genome. Orthologs are found throughout, but are limited to, the phylum Chordata (with a few exceptions in other phyla of the kingdom Animalia, like the Octopus bimaculoides).[21]
| Species | Common Name | NCBI Accession Number | Sequence Length (AA) | Millions of Years Since LCA[22] | % Identity | % Similarity |
|---|---|---|---|---|---|---|
| Homo sapiens | Human | NP_001093866.1 | 287 | - | - | - |
| Heterocephalus glaber | Naked mole rat | XP_004867342.1 | 235 | 90 | 62.2 | 68.2 |
| Mus musculus | Mouse | NP_001093864.1 | 233 | 90 | 54.9 | 62.8 |
| Fukomys damarensis | Damaraland mole-rat | XP_010614136.2 | 288 | 90 | 75.0 | 83.7 |
| Pteropus vampyrus | Large Flying Fox | XP_011362281.1 | 291 | 96 | 77.7 | 82.8 |
| Eptesicus fuscus | Big Brown Bat | XP_008160678.1 | 321 | 96 | 61.8 | 67.6 |
| Rhinolophus sinicus | Chinese Rufous Horseshoe Bat | XP_019575083.1 | 301 | 96 | 71.4 | 79.4 |
| Erinaceus europaeus | European Hedgehog | XP_007528011.1 | 284 | 96 | 63.8 | 71.4 |
| Condylura cristata | Star nosed mole | XP_012586937.1 | 291 | 96 | 69.8 | 79.0 |
| Camelus ferus | Wild Bactrian camel | XP_006174505.1 | 291 | 96 | 75.6 | 83.2 |
| Capra hircus | Goat | XP_013823176.1 | 285 | 96 | 73.1 | 77.9 |
| Bos taurus | Cattle | NP_001094753.1 | 290 | 96 | 75.5 | 81.0 |
| Panthera pardus | Leopard | XP_019277335.1 | 292 | 96 | 75.0 | 82.2 |
| Ursus maritimus | Polar Bear | XP_008698084.1 | 290 | 96 | 77.3 | 84.5 |
| Falco peregrinus | Peregrine Falcon | XP_013152712.1 | 231 | 312 | 36.2 | 44.6 |
| Apteryx mantelli | North Island Brown Kiwi | XP_013805202.1 | 197 | 312 | 36.9 | 41.9 |
| Python bivittatus | Burmese Python | XP_007425859.1 | 314 | 312 | 30.8 | 45.0 |
| Anolis carolinensis | Carolina anole | XP_003216202.2 | 320 | 312 | 35.3 | 42.7 |
| Xenopus laevis | African Clawed Frog | XP_018118010.1 | 307 | 352 | 36.9 | 52.2 |
| Nanorana parkeri | Frog | XP_018419829.1 | 307 | 352 | 36.4 | 45.8 |
| Callorhinchus milii | Australian Ghostshark | XP_007890694.1 | 293 | 473 | 28.0 | 34.9 |
| Ciona intestinalis | Sea squirt | XP_002125895.1 | 235 | 676 | 22.4 | 34.5 |
| Octopus bimaculoides | California two-spot octopus | XP_014784430.1 | 242 | 797 | 22.4 | 30.0 |
| Saccoglossus kowalevskii | Acorn Worm | XP_002735239.2 | 232 | 684 | 17.9 | 27.9 |
References
- GRCh38: Ensembl release 89: ENSG00000177994 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000040919 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Homo sapiens chromosome 2 open reading frame 73 (C2orf73), mRNA - Nucleotide - NCBI". www.ncbi.nlm.nih.gov. Retrieved 28 April 2017.
- "C2orf73 chromosome 2 open reading frame 73 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 28 April 2017.
- "C2orf73 Gene - GeneCards | CB073 Protein | CB073 Antibody". www.genecards.org. Retrieved 28 April 2017.
- "C2orf73 chromosome 2 open reading frame 73 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 28 April 2017.
- "SDSC Biology Workbench". workbench.sdsc.edu. Retrieved 28 April 2017.
- "I-TASSER server for protein structure and function prediction". zhanglab.ccmb.med.umich.edu. Retrieved 28 April 2017.
- Zhang Y (January 2008). "I-TASSER server for protein 3D structure prediction". BMC Bioinformatics. 9 (1): 40. doi:10.1186/1471-2105-9-40. PMC 2245901. PMID 18215316.
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (January 2015). "The I-TASSER Suite: protein structure and function prediction". Nature Methods. 12 (1): 7–8. doi:10.1038/nmeth.3213. PMC 4428668. PMID 25549265.
- Roy A, Kucukural A, Zhang Y (April 2010). "I-TASSER: a unified platform for automated protein structure and function prediction". Nature Protocols. 5 (4): 725–38. doi:10.1038/nprot.2010.5. PMC 2849174. PMID 20360767.
- "SDSC Biology Workbench". workbench.sdsc.edu. Retrieved 28 April 2017.
- "ExPASy: SIB Bioinformatics Resource Portal - Categories". www.expasy.org. Retrieved 28 April 2017.
- "PSORT II Prediction". psort.hgc.jp. Retrieved 28 April 2017.
- Hay RT (April 2005). "SUMO: a history of modification". Molecular Cell. 18 (1): 1–12. doi:10.1016/j.molcel.2005.03.012. PMID 15808504.
- "Home - GEO - NCBI". www.ncbi.nlm.nih.gov. Retrieved 28 April 2017.
- "Genomatix - NGS Data Analysis & Personalized Medicine". www.genomatix.de. Retrieved 28 April 2017.
- "PSICQUIC View". www.ebi.ac.uk. Retrieved 28 April 2017.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (September 1997). "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs". Nucleic Acids Research. 25 (17): 3389–402. doi:10.1093/nar/25.17.3389. PMC 146917. PMID 9254694.
- "TimeTree :: The Timescale of Life". www.timetree.org. Retrieved 28 April 2017.