Secoviridae
Secoviridae is a family of viruses in the order Picornavirales. Plants serve as natural hosts. There are currently 86 species in this family, divided among 8 genera or not assigned to a genus.[1][2][3][4] The family was created in 2009 with the grouping of families Sequiviridae, now dissolved, and Comoviridae, now subfamily Comovirinae, along with the then unassigned genera Cheravirus, Sadwavirus, and Torradovirus.[4]
| Secoviridae | |
|---|---|
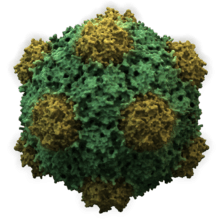 | |
| Structure of the Cowpea Mosaic Virus based on PDB 2BFU | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Picornavirales |
| Family: | Secoviridae |
| Genera | |
|
Subfamily: Comovirinae | |
Structure
Viruses in Secoviridae are non-enveloped, with icosahedral geometries, and T=pseudo3 symmetry. The diameter is around 25-30 nm. Genomes are linear and segmented, bipartite, around 24-7kb in length.[1][2][3]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Sequivirus | Icosahedral | Pseudo T=3 | Non-enveloped | Linear | Monopartite |
| Sadwavirus | Icosahedral | Pseudo T=3 | Non-enveloped | Linear | Segmented |
| Nepovirus | Icosahedral | Pseudo T=3 | Non-enveloped | Linear | Segmented |
| Fabavirus | Icosahedral | Pseudo T=3 | Non-enveloped | Linear | Segmented |
| Comovirus | Icosahedral | Pseudo T=3 | Non-enveloped | Linear | Segmented |
| Cheravirus | Icosahedral | Pseudo T=3 | Non-enveloped | Linear | Segmented |
| Torradovirus | Icosahedral | Pseudo T=3 | Non-enveloped | Linear | Segmented |
| Waikavirus | Icosahedral | Pseudo T=3 | Non-enveloped | Linear | Monopartite |
Life cycle
Viral replication is cytoplasmic. Entry into the host cell is achieved by penetration into the host cell. Replication follows the positive stranded RNA virus replication model. Positive stranded RNA virus transcription is the method of transcription. The virus exits the host cell by tubule-guided viral movement. Plants serve as the natural host. Transmission routes are mechanical.[1][2][3]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Sequivirus | Plants | None | Viral movement; mechanical inoculation | Viral movement | Cytoplasm | Cytoplasm | Mechanical inoculation: aphids; Mechanical inoculation: Cavariella aegopodii; Mechanical inoculation: Cavariella pastinacae |
| Sadwavirus | Plants | None | Viral movement; mechanical inoculation | Viral movement | Cytoplasm | Cytoplasm | Nematodes; mites; thrips |
| Nepovirus | Plants | None | Viral movement; mechanical inoculation | Viral movement | Cytoplasm | Cytoplasm | Nematodes; mites; thrips |
| Fabavirus | Plants | None | Viral movement; mechanical inoculation | Viral movement | Cytoplasm | Cytoplasm | Mechanical inoculation: aphids |
| Comovirus | Plants | None | Viral movement; mechanical inoculation | Viral movement | Cytoplasm | Cytoplasm | Mechanical inoculation: beetles |
| Cheravirus | Plants | None | Viral movement; mechanical inoculation | Viral movement | Cytoplasm | Cytoplasm | Nematodes; mites; thrips |
| Torradovirus | Plants | None | Viral movement; mechanical inoculation | Viral movement | Cytoplasm | Cytoplasm | Unknown |
| Waikavirus | Gramineae | Phloem; phloem parenchyma; bundle sheath | Viral movement | Viral movement | Cytoplasm | Cytoplasm | Mechanical innocuation: aphids; mechanical innocuation: leafhoppers |
Taxonomy
Group: ssRNA(+)
- Family: Secoviridae
- Sub-Family: Comovirinae
- Genus: Comovirus
- Andean potato mottle virus
- Bean pod mottle virus
- Bean rugose mosaic virus
- Broad bean stain virus
- Broad bean true mosaic virus
- Cowpea mosaic virus
- Cowpea severe mosaic virus
- Glycine mosaic virus
- Pea green mottle virus
- Pea mild mosaic virus
- Quail pea mosaic virus
- Radish mosaic virus
- Red clover mottle virus
- Squash mosaic virus
- Ullucus virus C
- Genus: Fabavirus
- Broad bean wilt virus 1
- Broad bean wilt virus 2
- Cucurbit mild mosaic virus
- Gentian mosaic virus
- Grapevine fabavirus
- Lamium mild mosaic virus
- Prunus virus F
- Genus: Nepovirus
- Aeonium ringspot virus
- Apricot latent ringspot virus
- Arabis mosaic virus
- Arracacha virus A
- Artichoke Aegean ringspot virus
- Artichoke Italian latent virus
- Artichoke yellow ringspot virus
- Beet ringspot virus
- Blackcurrant reversion virus
- Blueberry latent spherical virus
- Blueberry leaf mottle virus
- Cassava American latent virus
- Cassava green mottle virus
- Cherry leaf roll virus
- Chicory yellow mottle virus
- Cocoa necrosis virus
- Crimson clover latent virus
- Cycas necrotic stunt virus
- Grapevine Anatolian ringspot virus
- Grapevine Bulgarian latent virus
- Grapevine chrome mosaic virus
- Grapevine deformation virus
- Grapevine fanleaf virus
- Grapevine Tunisian ringspot virus
- Hibiscus latent ringspot virus
- Lucerne Australian latent virus
- Melon mild mottle virus
- Mulberry mosaic leaf roll associated virus
- Mulberry ringspot virus
- Myrobalan latent ringspot virus
- Olive latent ringspot virus
- Peach rosette mosaic virus
- Potato black ringspot virus
- Potato virus B
- Potato virus U
- Raspberry ringspot virus
- Soybean latent spherical virus
- Tobacco ringspot virus
- Tomato black ring virus
- Tomato ringspot virus
- Sub-Family: Unassigned
- Genus: Cheravirus
- Apple latent spherical virus
- Arracacha virus B
- Cherry rasp leaf virus
- Currant latent virus
- Stocky prune virus
- Genus: Sadwavirus
- Black raspberry necrosis virus
- Chocolate lily virus A
- Dioscorea mosaic associated virus
- Satsuma dwarf virus
- Strawberry mottle virus
- Genus: Sequivirus
- Carrot necrotic dieback virus
- Dandelion yellow mosaic virus
- Parsnip yellow fleck virus
- Genus: Torradovirus
- Carrot torradovirus 1
- Lettuce necrotic leaf curl virus
- Motherwort yellow mottle virus
- Squash chlorotic leaf spot virus
- Tomato marchitez virus
- Tomato torrado virus
- Genus: Unassigned
- Genus: Waikavirus
- Anthriscus yellows virus
- Bellflower vein chlorosis virus
- Maize chlorotic dwarf virus
- Rice tungro spherical virus
Evolution
The subfamily Comovirinae evolved ~1,000 years ago with extant species diversifying between 50 and 250 years ago.[5] This time period coincides with the intensification of agricultural practices in industrial societies.
The mutation rate has been estimated to be 9.29×10−3 to 2.74×10−3 subs/site/year.
References
- Thompson, JR; Dasgupta, I; Fuchs, M; Iwanami, T; Karasev, AV; Petrzik, K; Sanfaçon, H; Tzanetakis, I; van der Vlugt, R; Wetzel, T; Yoshikawa, N; ICTV Report Consortium (April 2017). "ICTV Virus Taxonomy Profile: Secoviridae". The Journal of General Virology. 98 (4): 529–531. doi:10.1099/jgv.0.000779. PMC 5657025. PMID 28452295.
- "Secoviridae". ICTV Online (10th) Report.
- "Viral Zone". ExPASy. Retrieved 15 June 2015.
- "Taxonomy". International Committee on Taxonomy of Viruses. Retrieved 25 March 2020.
- Thompson, JR; Kamath, N; Perry, KL (2014). "An evolutionary analysis of the secoviridae family of viruses". PLOS ONE. 9 (9): e106305. doi:10.1371/journal.pone.0106305. PMC 4152289. PMID 25180860.
External links
- ICTV Online (10th) Report: Secoviridae
- Viralzone: Secoviridae
- UniProt Taxonomy