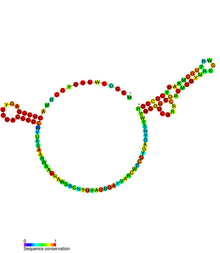Ribosomal frameshift
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA.[1] The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure.[2] It has been described mainly in viruses (especially retroviruses), retrotransposons and bacterial insertion elements, and also in some cellular genes.[3]
Process overview
Proteins are translated by reading tri-nucleotides on the mRNA strand, also known as codons, from one end of the mRNA to the other (from the 5' to the 3' end). Each codon is translated into a single amino acid. Therefore, a shift of any number of nucleotides that is not divisible by 3 in the reading frame will result in subsequent codons to be read differently.[4] This effectively changes the ribosomal reading frame.
Sentence example
In this example, the following sentence with three-letter words makes sense when read from the beginning:
|Start|THE CAT AND THE MAN ARE FAT ... |Start|123 123 123 123 123 123 123 ...
However, if the reading frame is shifted by one letter to between the T and H of the first word (effectively a +1 frameshift when considering the 0 position to be the initial position of T),
T|Start|HEC ATA NDT HEM ANA REF AT... -|Start|123 123 123 123 123 123 12...
then the sentence reads differently, making no sense.
DNA example
In this example, the following sequence is a region of the human mitochondrial genome with the two overlapping genes MT-ATP8 and MT-ATP6. When read from the beginning, these codons make sense to a ribosome and can be translated into amino acids (AA) under the vertebrate mitochondrial code:
|Start|AAC GAA AAT CTG TTC GCT TCA ... |Start|123 123 123 123 123 123 123 ... | AA | N E N L F A S ...
However, let's change the reading frame by starting one nucleotide downstream (effectively a "+1 frameshift" when considering the 0 position to be the initial position of A):
A|Start|ACG AAA ATC TGT TCG CTT CA... -|Start|123 123 123 123 123 123 12... | AA | T K I C S L ...
Now, because of this +1 frameshifting, the DNA sequence is read differently. The different codon reading frame therefore yields different amino acids.
In the case of a translating ribosome, a frameshift can either result in nonsense (a premature stop codon) after the frameshift, or the creation of a completely new protein after the frameshift. In the case where a frameshift results in nonsense, the NMD (nonsense-mediated mRNA decay) pathway may destroy the mRNA transcript, so frameshifting would serve as a method of regulating the expression level of the associated gene.[5]
Function
In viruses this phenomenon may be programmed to occur at particular sites and allows the virus to encode multiple types of proteins from the same mRNA. Notable examples include HIV-1 (human immunodeficiency virus),[6] RSV (Rous sarcoma virus)[7] and the influenza virus (flu),[8] which all rely on frameshifting to create a proper ratio of 0-frame (normal translation) and "trans-frame" (encoded by frameshifted sequence) proteins. Its use in viruses is primarily for compacting more genetic information into a shorter amount of genetic material.
In eukaryotes it appears to play a role in regulating gene expression levels by generating premature stops and producing nonfunctional transcripts.[3][9]
Types of frameshifting
The most common type of frameshifting is −1 frameshifting or programmed −1 ribosomal frameshifting (−1 PRF). Other, rarer types of frameshifting include +1 and −2 frameshifting.[2] −1 and +1 frameshifting are believed to be controlled by different mechanisms, which are discussed below. Both mechanisms are kinetically driven.
Programmed −1 ribosomal frameshifting
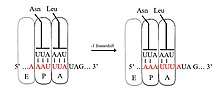
In −1 frameshifting, the ribosome slips back one nucleotide and continues translation in the −1 frame. There are typically three elements that comprise a −1 frameshift signal: a slippery sequence, a spacer region, and an RNA secondary structure. The slippery sequence fits a X_XXY_YYZ motif, where XXX is any three identical nucleotides (though some exceptions occur), YYY typically represents UUU or AAA, and Z is A, C or U. Because the structure of this motif contains 2 adjacent 3-nucleotide repeats it is believed that −1 frameshifting is described by a tandem slippage model, in which the ribosomal P-site tRNA anticodon re-pairs from XXY to XXX and the A-site anticodon re-pairs from YYZ to YYY simultaneously. These new pairings are identical to the 0-frame pairings except at their third positions. This difference does not significantly disfavor anticodon binding because the third nucleotide in a codon, known as the wobble position, has weaker tRNA anticodon binding specificity than the first and second nucleotides.[2][10] In this model, the motif structure is explained by the fact that the first and second positions of the anticodons must be able to pair perfectly in both the 0 and −1 frames. Therefore, nucleotides 2 and 1 must be identical, and nucleotides 3 and 2 must also be identical, leading to a required sequence of 3 identical nucleotides for each tRNA that slips.[11]
+1 ribosomal frameshifting
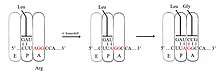
The slippery sequence for a +1 frameshift signal does not have the same motif, and instead appears to function by pausing the ribosome at a sequence encoding a rare amino acid.[12] Ribosomes do not translate proteins at a steady rate, regardless of the sequence. Certain codons take longer to translate, because there are not equal amounts of tRNA of that particular codon in the cytosol.[13] Due to this lag, there exist in small sections of codons sequences that control the rate of ribosomal frameshifting. Specifically, the ribosome must pause to wait for the arrival of a rare tRNA, and this increases the kinetic favorability of the ribosome and its associated tRNA slipping into the new frame.[12][14] In this model, the change in reading frame is caused by a single tRNA slip rather than two.
Controlling mechanisms
Ribosomal frameshifting may be controlled by mechanisms found in the mRNA sequence (cis-acting). This generally refers to a slippery sequence, a RNA secondary structure, or both. A −1 frameshift signal consists of both elements separated by a spacer region typically 5–9 nucleotides long.[2] Frameshifting may also be induced by other molecules which interact with the ribosome or the mRNA (trans-acting).
Frameshift signal elements
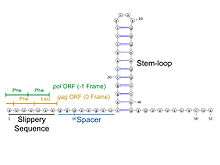
Slippery sequence
Slippery sequences can potentially make the reading ribosome "slip" and skip a number of nucleotides (usually only 1) and read a completely different frame thereafter. In programmed −1 ribosomal frameshifting, the slippery sequence fits a X_XXY_YYZ motif, where XXX is any three identical nucleotides (though some exceptions occur), YYY typically represents UUU or AAA, and Z is A, C or U. In the case of +1 frameshifting, the slippery sequence contains codons for which the corresponding tRNA is more rare, and the frameshift is favored because the codon in the new frame has a more common associated tRNA.[12] One example of a slippery sequence is the polyA on mRNA, which is known to induce ribosome slippage even in the absence of any other elements.[15]
RNA secondary structure
Efficient ribosomal frameshifting generally requires the presence of an RNA secondary structure to enhance the effects of the slippery sequence.[11] The RNA structure (which can be a stem-loop or pseudoknot) is thought to pause the ribosome on the slippery site during translation, forcing it to relocate and continue replication from the −1 position. It is believed that this occurs because the structure physically blocks movement of the ribosome by becoming stuck in the ribosome mRNA tunnel.[2] This model is supported by the fact that strength of the pseudoknot has been positively correlated with the level of frameshifting for associated mRNA.[3][16]
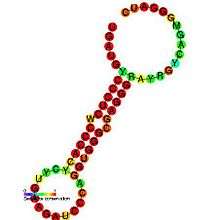
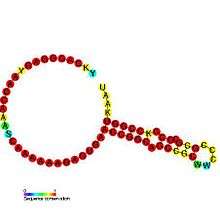
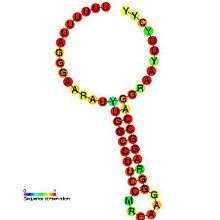
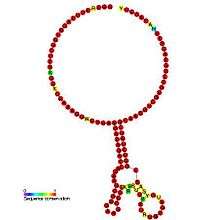
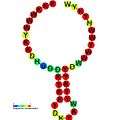
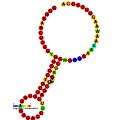
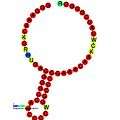
Below are examples of predicted secondary structures for frameshift elements shown to stimulate frameshifting in a variety of organisms. The majority of the structures shown are stem-loops, with the exception of the ALIL (apical loop-internal loop) pseudoknot structure. In these images, the larger and incomplete circles of mRNA represent linear regions. The secondary "stem-loop" structures, where "stems" are formed by a region of mRNA base pairing with another region on the same strand, are shown protruding from the linear DNA. The linear region of the HIV ribosomal frameshift signal contains a highly conserved UUU UUU A slippery sequence; many of the other predicted structures contain candidates for slippery sequences as well.
The mRNA sequences in the images can be read according to a set of guidelines. While A, T, C, and G represent a particular nucleotide at a position, there are also letters that represent ambiguity which are used when more than one kind of nucleotide could occur at that position. The rules of the International Union of Pure and Applied Chemistry (IUPAC) are as follows:[17]
| Symbol[17] | Description | Bases represented | Complement | ||||
|---|---|---|---|---|---|---|---|
| A | Adenine | A | 1 | T | |||
| C | Cytosine | C | G | ||||
| G | Guanine | G | C | ||||
| T | Thymine | T | A | ||||
| U | Uracil | U | A | ||||
| W | Weak | A | T | 2 | W | ||
| S | Strong | C | G | S | |||
| M | aMino | A | C | K | |||
| K | Keto | G | T | M | |||
| R | puRine | A | G | Y | |||
| Y | pYrimidine | C | T | R | |||
| B | not A (B comes after A) | C | G | T | 3 | V | |
| D | not C (D comes after C) | A | G | T | H | ||
| H | not G (H comes after G) | A | C | T | D | ||
| V | not T (V comes after T and U) | A | C | G | B | ||
| N | any Nucleotide (not a gap) | A | C | G | T | 4 | N |
| Z | Zero | 0 | Z | ||||
These symbols are also valid for RNA, except with U (uracil) replacing T (thymine).[17]
| Type | Distribution | Ref. |
|---|---|---|
| ALIL pseudoknot | Bacteria | [18] |
| Antizyme RNA frameshifting stimulation element | Invertebrates | [19] |
| Coronavirus frameshifting stimulation element | Coronavirus | [20] |
| DnaX ribosomal frameshifting element | Eukaryota, bacteria | [21] |
| HIV ribosomal frameshift signal | Viruses | |
| Insertion sequence IS1222 ribosomal frameshifting element | Eukaryota, bacteria | |
| Ribosomal frameshift | Viruses |
Trans-acting elements
Small molecules, proteins, and nucleic acids have been found to stimulate levels of frameshifting. For example, the mechanism of a negative feedback loop in the polyamine synthesis pathway is based on polyamine levels stimulating an increase in +1 frameshifts, which results in production of an inhibitory enzyme. Certain proteins which are needed for codon recognition or which bind directly to the mRNA sequence have also been shown to modulate frameshifting levels. MicroRNA (miRNA) molecules may hybridize to a RNA secondary structure and affect its strength.[5]
See also
References
- Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV (September 2016). "Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use". Nucleic Acids Research. 44 (15): 7007–7078. doi:10.1093/nar/gkw530. PMC 5009743. PMID 27436286.
- Napthine S, Ling R, Finch LK, Jones JD, Bell S, Brierley I, Firth AE (June 2017). "Protein-directed ribosomal frameshifting temporally regulates gene expression". Nature Communications. 8: 15582. Bibcode:2017NatCo...815582N. doi:10.1038/ncomms15582. PMC 5472766. PMID 28593994.
- Ketteler R (2012). "On programmed ribosomal frameshifting: the alternative proteomes". Frontiers in Genetics. 3: 242. doi:10.3389/fgene.2012.00242. PMC 3500957. PMID 23181069.
- Ivanov IP, Atkins JF (2007). "Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation". Nucleic Acids Research. 35 (6): 1842–1858. doi:10.1093/nar/gkm035. PMC 1874602. PMID 17332016.
- Dever TE, Dinman JD, Green R (August 2018). "Translation Elongation and Recoding in Eukaryotes". Cold Spring Harbor Perspectives in Biology. 10 (8): a032649. doi:10.1101/cshperspect.a032649. PMC 6071482. PMID 29610120.
- Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE (January 1988). "Characterization of ribosomal frameshifting in HIV-1 gag-pol expression". Nature. 331 (6153): 280–283. Bibcode:1988Natur.331..280J. doi:10.1038/331280a0. PMID 2447506.
- Jacks T, Madhani HD, Masiarz FR, Varmus HE (November 1988). "Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region". Cell. 55 (3): 447–458. doi:10.1016/0092-8674(88)90031-1. PMC 7133365. PMID 2846182.
- Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P (July 2012). "An overlapping protein-coding region in influenza A virus segment 3 modulates the host response". Science. 337 (6091): 199–204. Bibcode:2012Sci...337..199J. doi:10.1126/science.1222213. PMC 3552242. PMID 22745253.
- Advani VM, Dinman JD (January 2016). "Reprogramming the genetic code: The emerging role of ribosomal frameshifting in regulating cellular gene expression". BioEssays. 38 (1): 21–26. doi:10.1002/bies.201500131. PMC 4749135. PMID 26661048.
- Crick FH (August 1966). "Codon—anticodon pairing: the wobble hypothesis". Journal of Molecular Biology. 19 (2): 548–555. doi:10.1016/S0022-2836(66)80022-0. PMID 5969078.
- Brierley I (August 1995). "Ribosomal frameshifting viral RNAs". The Journal of General Virology. 76 (Pt 8) (8): 1885–1892. doi:10.1099/0022-1317-76-8-1885. PMID 7636469.
- Harger JW, Meskauskas A, Dinman JD (September 2002). "An "integrated model" of programmed ribosomal frameshifting". Trends in Biochemical Sciences. 27 (9): 448–454. doi:10.1016/S0968-0004(02)02149-7. PMID 12217519.
- Gurvich OL, Baranov PV, Gesteland RF, Atkins JF (June 2005). "Expression levels influence ribosomal frameshifting at the tandem rare arginine codons AGG_AGG and AGA_AGA in Escherichia coli". Journal of Bacteriology. 187 (12): 4023–4032. doi:10.1128/JB.187.12.4023-4032.2005. PMC 1151738. PMID 15937165.
- Caliskan N, Katunin VI, Belardinelli R, Peske F, Rodnina MV (June 2014). "Programmed −1 frameshifting by kinetic partitioning during impeded translocation". Cell. 157 (7): 1619–1631. doi:10.1016/j.cell.2014.04.041. PMID 24949973.
- Arthur L, Pavlovic-Djuranovic S, Smith-Koutmou K, Green R, Szczesny P, Djuranovic S (July 2015). "Translational control by lysine-encoding A-rich sequences". Science Advances. 1 (6): e1500154. Bibcode:2015SciA....1E0154A. doi:10.1126/sciadv.1500154. PMC 4552401. PMID 26322332.
- Hansen TM, Reihani SN, Oddershede LB, Sørensen MA (April 2007). "Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting". Proceedings of the National Academy of Sciences of the United States of America. 104 (14): 5830–5835. Bibcode:2007PNAS..104.5830H. doi:10.1073/pnas.0608668104. PMC 1838403. PMID 17389398.
- Nomenclature Committee of the International Union of Biochemistry (NC-IUB) (1984). "Nomenclature for Incompletely Specified Bases in Nucleic Acid Sequences". Retrieved 4 February 2008.
- Mazauric MH, Licznar P, Prère MF, Canal I, Fayet O (July 2008). "Apical loop-internal loop RNA pseudoknots: a new type of stimulator of −1 translational frameshifting in bacteria". The Journal of Biological Chemistry. 283 (29): 20421–20432. doi:10.1074/jbc.M802829200. PMID 18474594.
- Ivanov IP, Anderson CB, Gesteland RF, Atkins JF (June 2004). "Identification of a new antizyme mRNA +1 frameshifting stimulatory pseudoknot in a subset of diverse invertebrates and its apparent absence in intermediate species". Journal of Molecular Biology. 339 (3): 495–504. doi:10.1016/j.jmb.2004.03.082. PMC 7125782. PMID 15147837.
- Baranov PV, Henderson CM, Anderson CB, Gesteland RF, Atkins JF, Howard MT (February 2005). "Programmed ribosomal frameshifting in decoding the SARS-CoV genome". Virology. 332 (2): 498–510. doi:10.1016/j.virol.2004.11.038. PMID 15680415.
- Larsen B, Gesteland RF, Atkins JF (August 1997). "Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50%". Journal of Molecular Biology. 271 (1): 47–60. doi:10.1006/jmbi.1997.1162. PMC 7126992. PMID 9300054.
External links
- Frameshifting,+Ribosomal at the US National Library of Medicine Medical Subject Headings (MeSH)
- Wise2 — aligns a protein against a DNA sequence allowing frameshifts and introns
- FastY — compare a DNA sequence to a protein sequence database, allowing gaps and frameshifts
- Path — tool that compares two frameshift proteins (back-translation principle)
- Recode2 — Database of recoded genes, including those that require programmed Translational frameshift.