HIV ribosomal frameshift signal
HIV ribosomal frameshift signal is a ribosomal frameshift (PRF) that human immunodeficiency virus (HIV) uses to translate several different proteins from the same sequence.
| HIV-1 Ribosomal frameshift signal | |
|---|---|
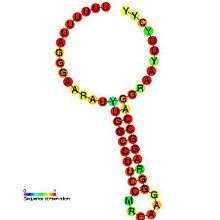 Predicted secondary structure and sequence conservation of HIV_FE | |
| Identifiers | |
| Symbol | HIV_FE |
| Rfam | RF00480 |
| Other data | |
| RNA type | Cis-reg; frameshift_element |
| Domain(s) | Viruses |
| SO | 0000233 |
| PDB structures | PDBe |
Intact and consistent protein biosynthesis relies on the ability of the ribosome to stay in the correct open reading frame (ORF) during translation.[1] When the ribosome fails to maintain the proper ORF, translation usually results in either incorrect protein synthesis or early termination as a result of the introduction of a premature stop codon.[2] However, a shift in the ORF is not universally deleterious, as many viruses capitalize on this phenomenon by using a programmed ribosomal frameshift (PRF) to translate several proteins from the same sequence, thereby maximizing the storage capacity of their genome.[2] Thus, many viruses (including HIV-1) are categorized as having a polycistronic genome, meaning they employ multiple active ORF's in a single gene.[2]
The HIV-1 virus requires a programmed -1 ribosomal frameshift signal (the HIV-1 Ribosomal Frameshift Signal) for the expression of the Pol gene, which is an example of a cis-acting element of gene regulation. In HIV-1, the gag ORF that encodes the 55 kDa Gag protein, the major viral structural protein, is located at the 5' end of the full-length viral mRNA.[3] Translation of the 160 kDa Gag-Pol polyprotein is contingent on a -1 ribosomal frameshift event revealing the pol ORF.[4] The pol ORF is located 3' to the gag ORF and encodes the Pol polyprotein, which is eventually cleaved into the viral enzymatic proteins (protease, reverse transcriptase, and integrase).
As a result, the HIV-1 ribosomal frameshift signal is highly regulated, as it modulates the expression levels of the Gag protein relative to the Gag-Pol polyprotein. The efficiency of the HIV-1 ribosomal frameshift signal determines the ratio of the Gag to Gag-Pol proteins synthesized, with a frameshift event occurring in approximately 5% of the total translation events, resulting in a roughly 20:1 Gag/Gag-Pol ratio.[1] Preservation of this ratio has been shown to be essential to HIV-1 infectivity and structure, as even small changes in the efficiency of the frameshift lead to inhibition of viral propagation.[3] The dependence of the HIV-1 virus on this ribosomal frameshift signal has generated interest in the frameshift as a target for novel antiviral therapeutics.[4][5]
Structure and mechanism
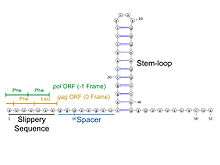
The HIV-1 ribosomal frameshift signal requires two cis-acting elements: a heptameric "slippery site" and a downstream secondary RNA structure separated by an 8-nucleotide spacer.[3][4] The "Slippery Site" in HIV-1 is the heptamer 5'-U UUU UUA-3' (gag ORF indicated by the spaces), where frameshifting occurs.[3][4] This heptamer is inherently "slippery", as data has shown that even in the absence of the downstream secondary RNA structure, frameshifting still occurs at roughly 0.0001% to 0.1% per codon.[2] It is generally accepted that the downstream secondary RNA structure exists as a stem-loop structure as shown below. However, there is also evidence that the frameshift signal may exist as a pseudoknot structure or as an intramolecular RNA triplex.[2][4] Regardless of the exact conformation of the downstream secondary RNA structure, it is believed that the structure leads to the translocating ribosome stalling over the slippery site, increasing the probability of a -1 ribosomal frameshift to reveal the pol ORF (5'-UUU UUU A-3'), bypassing a downstream stop codon present in the gag ORF and allowing the Gag-Pol polyprotein to be translated.[3][5] Data has shown that the 8-nucleotide spacer is essential to the programmed ribosomal frameshift as well, as deletions within the spacer region decrease the stability of the downstream secondary RNA structure, thereby affecting the ability of the HIV-1 ribosomal frameshift signal to induce a -1 frameshift.[2]
Modulators
Endogenous cellular factors may also modulate the HIV-1 ribosomal frameshift signal, as it has been reported that the eukaryotic release factor eRF1 plays a role in programmed ribosomal frameshift in HIV-1, as decreased levels of eRF1 lead to an increase in programmed ribosomal frameshift in HIV-1.[1] However, because eRF1 is known to complex with at least 32 cellular binding partners, it remains unclear if eRF1 acts independently to modulate PRF in HIV-1 or if it is part of a larger regulatory protein complex.[1]
As a potential therapeutic target
The HIV-1 ribosomal frameshift signal has emerged as a potential therapeutic target for the HIV-1 virus due to the requirement of the programmed ribosomal frameshift for the regulation of the Gag/Gag-Pol protein ratio and the relatively conserved structure.[4] Additionally, because the HIV-1 ribosomal frameshift signal relies on interactions between the viral mRNA and the host translational machinery, it is likely a more stable therapeutic target, because any selective pressure caused by a therapeutic compound would have to occur on the evolutionary time scale of the host instead of the rapidly evolving HIV-1 virus.[4] As a result, this may also reduce the risk of drug-resistant mutants experienced by other HIV-1 antiretroviral therapies.[4]
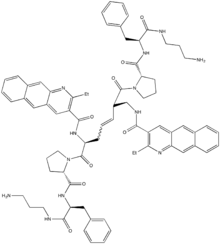
Recently (January 2014), the first therapeutic compound targeted at the HIV-1 ribosomal frameshift signal was reported by Ofori et al.[5] The lead compound was developed from a "hit" compound discovered through a resin-bound dynamic combinatorial library screen, and the structure is shown at right.[5] The EC50 values were reported to be 3.9uM for the Z conformation and 25.6uM for the E conformation. The lead compound is symmetrical whereas the target downstream secondary RNA structure is non-symmetrical, suggesting that both supposed intercalators are necessary for high-affinity binding.[5] Using a dual-luciferase assay, they concluded that the compound functions by enhancing the frameshifting efficiency of the HIV-1 ribosomal frameshift signal, resulting in a decrease in the Gag/Gag-Pol protein ratio and thereby preventing the proper maturation of the viral particle and ultimately inhibiting infection.[5] Moving forward, structural studies of the interactions between the lead compound and the downstream secondary RNA structure of the HIV-1 ribosomal frameshift signal will be vital to understanding the reason for high affinity and the method of action.[5]
References
- Kobayashi Y, Zhuang J, Peltz S, Dougherty J (June 2010). "Identification of a cellular factor that modulates HIV-1 programmed ribosomal frameshifting". The Journal of Biological Chemistry. 285 (26): 19776–19784. doi:10.1074/jbc.M109.085621. PMC 2888388. PMID 20418372.
- Mouzakis KD, Lang AL, Vander Meulen KA, Easterday PD, Butcher SE (February 2013). "HIV-1 frameshift efficiency is primarily determined by the stability of base pairs positioned at the mRNA entrance channel of the ribosome". Nucleic Acids Research. 41 (3): 1901–1913. doi:10.1093/nar/gks1254. PMC 3561942. PMID 23248007.
- Dinman JD, Richter S, Plant EP, Taylor RC, Hammell AB, Rana TM (April 2002). "The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure". Proceedings of the National Academy of Sciences of the United States of America. 99 (8): 5331–5336. doi:10.1073/pnas.082102199. PMC 122769. PMID 11959986.
- Biswas P, Jiang X, Pacchia AL, Dougherty JP, Peltz SW (February 2004). "The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy". Journal of Virology. 78 (4): 2082–2087. doi:10.1128/jvi.78.4.2082-2087.2004. PMC 369415. PMID 14747573.
- Ofori LO, Hilimire TA, Bennett RP, Brown NW, Smith HC, Miller BL (February 2014). "High-affinity recognition of HIV-1 frameshift-stimulating RNA alters frameshifting in vitro and interferes with HIV-1 infectivity". Journal of Medicinal Chemistry. 57 (3): 723–732. doi:10.1021/jm401438g. PMC 3954503. PMID 24387306.