PSMC5
26S protease regulatory subunit 8, also known as 26S proteasome AAA-ATPase subunit Rpt6, is an enzyme that in humans is encoded by the PSMC5 gene.[5][6][7] This protein is one of the 19 essential subunits of a complete assembled 19S proteasome complex[8] Six 26S proteasome AAA-ATPase subunits (Rpt1, Rpt2, Rpt3, Rpt4, Rpt5, and Rpt6 (this protein)) together with four non-ATPase subunits (Rpn1, Rpn2, Rpn10, and Rpn13) form the base sub complex of 19S regulatory particle for proteasome complex.[8]
Gene
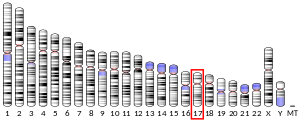
The gene PSMC5 encodes one of the ATPase subunits, a member of the triple-A family of ATPases which have a chaperone-like activity. In addition to participation in proteasome functions, this subunit may participate in transcriptional regulation since it has been shown to interact with the thyroid hormone receptor and retinoid X receptor-alpha.[7] The human PSMC5 gene has 13 exons and locates at chromosome band 17q23.3.
Protein
The human protein 26S protease regulatory subunit 8 is 45.6kDa in size and composed of 406 amino acids. The calculated theoretical pI of this protein is 8.23.[9]
Complex assembly
26S proteasome complex is usually consisted of a 20S core particle (CP, or 20S proteasome) and one or two 19S regulatory particles (RP, or 19S proteasome) on either one side or both side of the barrel-shaped 20S. The CP and RPs pertain distinct structural characteristics and biological functions. In brief, 20S sub complex presents three types proteolytic activities, including caspase-like, trypsin-like, and chymotrypsin-like activities. These proteolytic active sites located in the inner side of a chamber formed by 4 stacked rings of 20S subunits, preventing random protein-enzyme encounter and uncontrolled protein degradation. The 19S regulatory particles can recognize ubiquitin-labeled protein as degradation substrate, unfold the protein to linear, open the gate of 20S core particle, and guide the substate into the proteolytic chamber. To meet such functional complexity, 19S regulatory particle contains at least 18 constitutive subunits. These subunits can be categorized into two classes based on the ATP dependence of subunits, ATP-dependent subunits and ATP-independent subunits. According to the protein interaction and topological characteristics of this multisubunit complex, the 19S regulatory particle is composed of a base and a lid subcomplex. The base consists of a ring of six AAA ATPases (Subunit Rpt1-6, systematic nomenclature) and four non-ATPase subunits (Rpn1, Rpn2, Rpn10, and Rpn13). Thus, 26S protease regulatory subunit 4 (Rpt2) is an essential component of forming the base subcomplex of 19S regulatory particle. For the assembly of 19S base sub complex, four sets of pivotal assembly chaperons (Hsm3/S5b, Nas2/P27, Nas6/P28, and Rpn14/PAAF1, nomenclature in yeast/mammals) were identified by four groups independently.[10][11][12][13][14][15] These 19S regulatory particle base-dedicated chaperons all binds to individual ATPase subunits through the C-terminal regions. For example, Hsm3/S5b binds to the subunit Rpt1 and Rpt2 (this protein), Nas2/p27 to Rpt5, Nas6/p28 to Rpt3, and Rpn14/PAAAF1 to Rpt6 (this protein), respectively. Subsequently, three intermediate assembly modules are formed as following, the Nas6/p28-Rpt3-Rpt6-Rpn14/PAAF1 module, the Nas2/p27-Rpt4-Rpt5 module, and the Hsm3/S5b-Rpt1-Rpt2-Rpn2 module. Eventually, these three modules assemble together to form the heterohexameric ring of 6 Atlases with Rpn1. The final addition of Rpn13 indicates the completion of 19S base sub complex assembly.[8]
Function
As the degradation machinery that is responsible for ~70% of intracellular proteolysis,[16] proteasome complex (26S proteasome) plays a critical roles in maintaining the homeostasis of cellular proteome. Accordingly, misfolded proteins and damaged protein need to be continuously removed to recycle amino acids for new synthesis; in parallel, some key regulatory proteins fulfill their biological functions via selective degradation; furthermore, proteins are digested into peptides for MHC class I antigen presentation. To meet such complicated demands in biological process via spatial and temporal proteolysis, protein substrates have to be recognized, recruited, and eventually hydrolyzed in a well controlled fashion. Thus, 19S regulatory particle pertains a series of important capabilities to address these functional challenges. To recognize protein as designated substrate, 19S complex has subunits that are capable to recognize proteins with a special degradative tag, the ubiquitinylation. It also have subunits that can bind with nucleotides (e.g., ATPs) in order to facilitate the association between 19S and 20S particles, as well as to cause confirmation changes of alpha subunit C-terminals that form the substate entrance of 20S complex.
The ATPases subunits assemble into a six-membered ring with a sequence of Rpt1–Rpt5–Rpt4–Rpt3–Rpt6–Rpt2, which interacts with the seven-membered alpha ring of 20S core particle and eastablishs an asymmetric interface between the 19S RP and the 20S CP.[17][18] Three C-terminal tails with HbYX motifs of distinct Rpt ATPases insert into pockets between two defined alpha subunits of the CP and regulate the gate opening of the central channels in the CP alpha ring.[19][20] Evidence showed that ATPase subunit Rpt5, along with other ubuiqintinated 19S proteasome subunits (Rpn13, Rpn10) and the deubiquitinating enzyme Uch37, can be ubiquitinated in situ by proteasome-associating ubiquitination enzymes. Ubiquitination of proteasome subunits can regulates proteasomal activity in response to the alteration of cellular ubiquitination levels.[21]
Interactions
PSMC5 has been shown to interact with:
References
- GRCh38: Ensembl release 89: ENSG00000087191 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000020708 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Tanahashi N, Suzuki M, Fujiwara T, Takahashi E, Shimbara N, Chung CH, Tanaka K (March 1998). "Chromosomal localization and immunological analysis of a family of human 26S proteasomal ATPases". Biochem Biophys Res Commun. 243 (1): 229–32. doi:10.1006/bbrc.1997.7892. PMID 9473509.
- Hoyle J, Tan KH, Fisher EM (March 1997). "Localization of genes encoding two human one-domain members of the AAA family: PSMC5 (the thyroid hormone receptor-interacting protein, TRIP1) and PSMC3 (the Tat-binding protein, TBP1)". Hum Genet. 99 (2): 285–8. doi:10.1007/s004390050356. PMID 9048938.
- "Entrez Gene: PSMC5 proteasome (prosome, macropain) 26S subunit, ATPase, 5".
- Gu ZC, Enenkel C (Dec 2014). "Proteasome assembly". Cellular and Molecular Life Sciences. 71 (24): 4729–45. doi:10.1007/s00018-014-1699-8. PMID 25107634.
- "Uniprot: P62195 - PRS8_HUMAN".
- Le Tallec B, Barrault MB, Guérois R, Carré T, Peyroche A (Feb 2009). "Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome". Molecular Cell. 33 (3): 389–99. doi:10.1016/j.molcel.2009.01.010. PMID 19217412.
- Funakoshi M, Tomko RJ, Kobayashi H, Hochstrasser M (May 2009). "Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base". Cell. 137 (5): 887–99. doi:10.1016/j.cell.2009.04.061. PMC 2718848. PMID 19446322.
- Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D (Jun 2009). "Hexameric assembly of the proteasomal ATPases is templated through their C termini". Nature. 459 (7248): 866–70. Bibcode:2009Natur.459..866P. doi:10.1038/nature08065. PMC 2722381. PMID 19412160.
- Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, Finley D (Jun 2009). "Chaperone-mediated pathway of proteasome regulatory particle assembly". Nature. 459 (7248): 861–5. Bibcode:2009Natur.459..861R. doi:10.1038/nature08063. PMC 2727592. PMID 19412159.
- Saeki Y, Toh-E A, Kudo T, Kawamura H, Tanaka K (May 2009). "Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle". Cell. 137 (5): 900–13. doi:10.1016/j.cell.2009.05.005. PMID 19446323.
- Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T, Tanaka K, Murata S (May 2009). "Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones". Cell. 137 (5): 914–25. doi:10.1016/j.cell.2009.05.008. PMID 19490896.
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL (Sep 1994). "Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules". Cell. 78 (5): 761–71. doi:10.1016/s0092-8674(94)90462-6. PMID 8087844.
- Tian G, Park S, Lee MJ, Huck B, McAllister F, Hill CP, Gygi SP, Finley D (Nov 2011). "An asymmetric interface between the regulatory and core particles of the proteasome". Nature Structural & Molecular Biology. 18 (11): 1259–67. doi:10.1038/nsmb.2147. PMC 3210322. PMID 22037170.
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A (Feb 2012). "Complete subunit architecture of the proteasome regulatory particle". Nature. 482 (7384): 186–91. Bibcode:2012Natur.482..186L. doi:10.1038/nature10774. PMC 3285539. PMID 22237024.
- Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN (Nov 2008). "Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome". The Journal of Biological Chemistry. 283 (46): 31813–31822. doi:10.1074/jbc.M805935200. PMC 2581596. PMID 18796432.
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL (Sep 2007). "Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry". Molecular Cell. 27 (5): 731–744. doi:10.1016/j.molcel.2007.06.033. PMC 2083707. PMID 17803938.
- Jacobson AD, MacFadden A, Wu Z, Peng J, Liu CW (Jun 2014). "Autoregulation of the 26S proteasome by in situ ubiquitination". Molecular Biology of the Cell. 25 (12): 1824–35. doi:10.1091/mbc.E13-10-0585. PMC 4055262. PMID 24743594.
- Ishizuka T, Satoh T, Monden T, Shibusawa N, Hashida T, Yamada M, Mori M (August 2001). "Human immunodeficiency virus type 1 Tat binding protein-1 is a transcriptional coactivator specific for TR". Mol. Endocrinol. 15 (8): 1329–43. doi:10.1210/mend.15.8.0680. PMID 11463857.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514.
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D (2007). "Large-scale mapping of human protein-protein interactions by mass spectrometry". Mol. Syst. Biol. 3: 89. doi:10.1038/msb4100134. PMC 1847948. PMID 17353931.
- Su K, Yang X, Roos MD, Paterson AJ, Kudlow JE (June 2000). "Human Sug1/p45 is involved in the proteasome-dependent degradation of Sp1". Biochem. J. 348. 348 Pt 2 (2): 281–9. doi:10.1042/0264-6021:3480281. PMC 1221064. PMID 10816420.
- Wang YT, Chuang JY, Shen MR, Yang WB, Chang WC, Hung JJ (July 2008). "Sumoylation of specificity protein 1 augments its degradation by changing the localization and increasing the specificity protein 1 proteolytic process". J. Mol. Biol. 380 (5): 869–85. doi:10.1016/j.jmb.2008.05.043. PMID 18572193.
- Weeda G, Rossignol M, Fraser RA, Winkler GS, Vermeulen W, van 't Veer LJ, Ma L, Hoeijmakers JH, Egly JM (June 1997). "The XPB subunit of repair/transcription factor TFIIH directly interacts with SUG1, a subunit of the 26S proteasome and putative transcription factor". Nucleic Acids Res. 25 (12): 2274–83. doi:10.1093/nar/25.12.2274. PMC 146752. PMID 9173976.
Further reading
- Coux O, Tanaka K, Goldberg AL (1996). "Structure and functions of the 20S and 26S proteasomes". Annu. Rev. Biochem. 65 (1): 801–47. doi:10.1146/annurev.bi.65.070196.004101. PMID 8811196.
- Goff SP (2003). "Death by deamination: a novel host restriction system for HIV-1". Cell. 114 (3): 281–3. doi:10.1016/S0092-8674(03)00602-0. PMID 12914693.
- Nelbock P, Dillon PJ, Perkins A, Rosen CA (1990). "A cDNA for a protein that interacts with the human immunodeficiency virus Tat transactivator". Science. 248 (4963): 1650–3. Bibcode:1990Sci...248.1650N. doi:10.1126/science.2194290. PMID 2194290.
- Akiyama K, Yokota K, Kagawa S, Shimbara N, DeMartino GN, Slaughter CA, Noda C, Tanaka K (1995). "cDNA cloning of a new putative ATPase subunit p45 of the human 26S proteasome, a homolog of yeast transcriptional factor Sug1p". FEBS Lett. 363 (1–2): 151–6. doi:10.1016/0014-5793(95)00304-R. PMID 7729537.
- Lee JW, Choi HS, Gyuris J, Brent R, Moore DD (1995). "Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor". Mol. Endocrinol. 9 (2): 243–54. doi:10.1210/me.9.2.243. PMID 7776974.
- Lee JW, Ryan F, Swaffield JC, Johnston SA, Moore DD (1995). "Interaction of thyroid-hormone receptor with a conserved transcriptional mediator". Nature. 374 (6517): 91–4. Bibcode:1995Natur.374...91L. doi:10.1038/374091a0. PMID 7870181.
- Shaw DR, Ennis HL (1993). "Molecular cloning and developmental regulation of Dictyostelium discoideum homologues of the human and yeast HIV1 Tat-binding protein". Biochem. Biophys. Res. Commun. 193 (3): 1291–6. doi:10.1006/bbrc.1993.1765. PMID 8323548.
- Ohana B, Moore PA, Ruben SM, Southgate CD, Green MR, Rosen CA (1993). "The type 1 human immunodeficiency virus Tat binding protein is a transcriptional activator belonging to an additional family of evolutionarily conserved genes". Proc. Natl. Acad. Sci. U.S.A. 90 (1): 138–42. Bibcode:1993PNAS...90..138O. doi:10.1073/pnas.90.1.138. PMC 45615. PMID 8419915.
- Dubiel W, Ferrell K, Rechsteiner M (1993). "Peptide sequencing identifies MSS1, a modulator of HIV Tat-mediated transactivation, as subunit 7 of the 26 S protease". FEBS Lett. 323 (3): 276–8. doi:10.1016/0014-5793(93)81356-5. PMID 8500623.
- vom Baur E, Zechel C, Heery D, Heine MJ, Garnier JM, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R (1996). "Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1". EMBO J. 15 (1): 110–24. doi:10.1002/j.1460-2075.1996.tb00339.x. PMC 449923. PMID 8598193.
- Andersson B, Wentland MA, Ricafrente JY, Liu W, Gibbs RA (1996). "A "double adaptor" method for improved shotgun library construction". Anal. Biochem. 236 (1): 107–13. doi:10.1006/abio.1996.0138. PMID 8619474.
- Wang W, Chevray PM, Nathans D (1996). "Mammalian Sug1 and c-Fos in the nuclear 26S proteasome". Proc. Natl. Acad. Sci. U.S.A. 93 (16): 8236–40. Bibcode:1996PNAS...93.8236W. doi:10.1073/pnas.93.16.8236. PMC 38653. PMID 8710853.
- Seeger M, Ferrell K, Frank R, Dubiel W (1997). "HIV-1 tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation". J. Biol. Chem. 272 (13): 8145–8. doi:10.1074/jbc.272.13.8145. PMID 9079628.
- Yu W, Andersson B, Worley KC, Muzny DM, Ding Y, Liu W, Ricafrente JY, Wentland MA, Lennon G, Gibbs RA (1997). "Large-scale concatenation cDNA sequencing". Genome Res. 7 (4): 353–8. doi:10.1101/gr.7.4.353. PMC 139146. PMID 9110174.
- Weeda G, Rossignol M, Fraser RA, Winkler GS, Vermeulen W, van 't Veer LJ, Ma L, Hoeijmakers JH, Egly JM (1997). "The XPB subunit of repair/transcription factor TFIIH directly interacts with SUG1, a subunit of the 26S proteasome and putative transcription factor". Nucleic Acids Res. 25 (12): 2274–83. doi:10.1093/nar/25.12.2274. PMC 146752. PMID 9173976.
- Chen Y, Sharp ZD, Lee WH (1997). "HEC binds to the seventh regulatory subunit of the 26 S proteasome and modulates the proteolysis of mitotic cyclins". J. Biol. Chem. 272 (38): 24081–7. doi:10.1074/jbc.272.38.24081. PMID 9295362.
- Tipler CP, Hutchon SP, Hendil K, Tanaka K, Fishel S, Mayer RJ (1998). "Purification and characterization of 26S proteasomes from human and mouse spermatozoa". Mol. Hum. Reprod. 3 (12): 1053–60. doi:10.1093/molehr/3.12.1053. PMID 9464850.