Metabolomics
Metabolomics is the scientific study of chemical processes involving metabolites, the small molecule substrates, intermediates and products of metabolism. Specifically, metabolomics is the "systematic study of the unique chemical fingerprints that specific cellular processes leave behind", the study of their small-molecule metabolite profiles.[1] The metabolome represents the complete set of metabolites in a biological cell, tissue, organ or organism, which are the end products of cellular processes.[2] mRNA gene expression data and proteomic analyses reveal the set of gene products being produced in the cell, data that represents one aspect of cellular function. Conversely, metabolic profiling can give an instantaneous snapshot of the physiology of that cell, and thus, metabolomics provides a direct "functional readout of the physiological state" of an organism.[3] One of the challenges of systems biology and functional genomics is to integrate genomics, transcriptomic, proteomic, and metabolomic information to provide a better understanding of cellular biology.
History
The concept that individuals might have a "metabolic profile" that could be reflected in the makeup of their biological fluids was introduced by Roger Williams in the late 1940s,[4] who used paper chromatography to suggest characteristic metabolic patterns in urine and saliva were associated with diseases such as schizophrenia. However, it was only through technological advancements in the 1960s and 1970s that it became feasible to quantitatively (as opposed to qualitatively) measure metabolic profiles. [5] The term "metabolic profile" was introduced by Horning, et al. in 1971 after they demonstrated that gas chromatography-mass spectrometry (GC-MS) could be used to measure compounds present in human urine and tissue extracts.[6][7] The Horning group, along with that of Linus Pauling and Arthur B. Robinson led the development of GC-MS methods to monitor the metabolites present in urine through the 1970s.[8]
Concurrently, NMR spectroscopy, which was discovered in the 1940s, was also undergoing rapid advances. In 1974, Seeley et al. demonstrated the utility of using NMR to detect metabolites in unmodified biological samples.[9] This first study on muscle highlighted the value of NMR in that it was determined that 90% of cellular ATP is complexed with magnesium. As sensitivity has improved with the evolution of higher magnetic field strengths and magic angle spinning, NMR continues to be a leading analytical tool to investigate metabolism.[6][10] Recent efforts to utilize NMR for metabolomics have been largely driven by the laboratory of Jeremy K. Nicholson at Birkbeck College, University of London and later at Imperial College London. In 1984, Nicholson showed 1H NMR spectroscopy could potentially be used to diagnose diabetes mellitus, and later pioneered the application of pattern recognition methods to NMR spectroscopic data.[11][12]
In 1995 liquid chromatography mass spectrometry metabolomics experiments[13] were performed by Gary Siuzdak while working with Richard Lerner (then president of The Scripps Research Institute) and Benjamin Cravatt, to analyze the cerebral spinal fluid from sleep deprived animals. One molecule of particular interest, oleamide, was observed and later shown to have sleep inducing properties. This work is one of the earliest such experiments combining liquid chromatography and mass spectrometry in metabolomics.
In 2005, the first metabolomics tandem mass spectrometry database, METLIN,[14][15] for characterizing human metabolites was developed in the Siuzdak laboratory at The Scripps Research Institute. METLIN has since grown and as of July 1, 2019, METLIN contains over 450,000 metabolites and other chemical entities, each compound having experimental tandem mass spectrometry data generated from molecular standards at multiple collision energies and in positive and negative ionization modes. METLIN is the largest repository of tandem mass spectrometry data of its kind. 2005 was also the year in which the dedicated academic journal Metabolomics first appeared, founded by its current editor-in-chief Professor Roy Goodacre.
In 2005, the Siuzdak lab was engaged in identifying metabolites associated with sepsis and in an effort to address the issue of statistically identifying the most relevant dysregulated metabolites across hundreds of LC/MS datasets, the first algorithm was developed to allow for the nonlinear alignment of mass spectrometry metabolomics data. Called XCMS,[16] where the "X" constitutes any chromatographic technology, it has since (2012)[17] been developed as an online tool and as of 2019 (with METLIN) has over 30,000 registered users.
On 23 January 2007, the Human Metabolome Project, led by David Wishart of the University of Alberta, Canada, completed the first draft of the human metabolome, consisting of a database of approximately 2500 metabolites, 1200 drugs and 3500 food components.[18][19] Similar projects have been underway in several plant species, most notably Medicago truncatula[20] and Arabidopsis thaliana[21] for several years.
As late as mid-2010, metabolomics was still considered an "emerging field".[22] Further, it was noted that further progress in the field depended in large part, through addressing otherwise "irresolvable technical challenges", by technical evolution of mass spectrometry instrumentation.[22]
In 2015, real-time metabolome profiling was demonstrated for the first time.[23]
Metabolome

Metabolome refers to the complete set of small-molecule (<1.5 kDa)[24] metabolites (such as metabolic intermediates, hormones and other signaling molecules, and secondary metabolites) to be found within a biological sample, such as a single organism.[25][26] The word was coined in analogy with transcriptomics and proteomics; like the transcriptome and the proteome, the metabolome is dynamic, changing from second to second. Although the metabolome can be defined readily enough, it is not currently possible to analyse the entire range of metabolites by a single analytical method.
The first metabolite database (called METLIN) for searching fragmentation data from tandem mass spectrometry experiments was developed by the Siuzdak lab at The Scripps Research Institute in 2005.[14][15] METLIN contains over 450,000 metabolites and other chemical entities, each compound having experimental tandem mass spectrometry data. In 2006,[16] the Siuzdak lab also developed the first algorithm to allow for the nonlinear alignment of mass spectrometry metabolomics data. Called XCMS, where the "X" constitutes any chromatographic technology, it has since (2012)[17] been developed as an online tool and as of 2019 (with METLIN) has over 30,000 registered users.
In January 2007, scientists at the University of Alberta and the University of Calgary completed the first draft of the human metabolome. The Human Metabolome Database (HMDB) is perhaps the most extensive public metabolomic spectral database to date.[27] The HMDB stores more than 40,000 different metabolite entries. They catalogued approximately 2500 metabolites, 1200 drugs and 3500 food components that can be found in the human body, as reported in the literature.[18] This information, available at the Human Metabolome Database (www.hmdb.ca) and based on analysis of information available in the current scientific literature, is far from complete.[28] In contrast, much more is known about the metabolomes of other organisms. For example, over 50,000 metabolites have been characterized from the plant kingdom, and many thousands of metabolites have been identified and/or characterized from single plants.[29][30]
Each type of cell and tissue has a unique metabolic ‘fingerprint’ that can elucidate organ or tissue-specific information. Bio-specimens used for metabolomics analysis include but not limit to plasma, serum, urine, saliva, feces, muscle, sweat, exhaled breath and gastrointestinal fluid.[31] The ease of collection facilitates high temporal resolution, and because they are always at dynamic equilibrium with the body, they can describe the host as a whole.[32] Genome can tell what could happen, transcriptome can tell what appears to be happening, proteome can tell what makes it happen and metabolome can tell what has happened and what is happening.[33]
Metabolites
Metabolites are the substrates, intermediates and products of metabolism. Within the context of metabolomics, a metabolite is usually defined as any molecule less than 1.5 kDa in size.[24] However, there are exceptions to this depending on the sample and detection method. For example, macromolecules such as lipoproteins and albumin are reliably detected in NMR-based metabolomics studies of blood plasma.[34] In plant-based metabolomics, it is common to refer to "primary" and "secondary" metabolites. A primary metabolite is directly involved in the normal growth, development, and reproduction. A secondary metabolite is not directly involved in those processes, but usually has important ecological function. Examples include antibiotics and pigments.[35] By contrast, in human-based metabolomics, it is more common to describe metabolites as being either endogenous (produced by the host organism) or exogenous.[36] Metabolites of foreign substances such as drugs are termed xenometabolites.[37]
The metabolome forms a large network of metabolic reactions, where outputs from one enzymatic chemical reaction are inputs to other chemical reactions. Such systems have been described as hypercycles.
Metabonomics
Metabonomics is defined as "the quantitative measurement of the dynamic multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modification". The word origin is from the Greek μεταβολή meaning change and nomos meaning a rule set or set of laws.[38] This approach was pioneered by Jeremy Nicholson at Murdoch University and has been used in toxicology, disease diagnosis and a number of other fields. Historically, the metabonomics approach was one of the first methods to apply the scope of systems biology to studies of metabolism.[39][40][41]
There has been some disagreement over the exact differences between 'metabolomics' and 'metabonomics'. The difference between the two terms is not related to choice of analytical platform: although metabonomics is more associated with NMR spectroscopy and metabolomics with mass spectrometry-based techniques, this is simply because of usages amongst different groups that have popularized the different terms. While there is still no absolute agreement, there is a growing consensus that 'metabolomics' places a greater emphasis on metabolic profiling at a cellular or organ level and is primarily concerned with normal endogenous metabolism. 'Metabonomics' extends metabolic profiling to include information about perturbations of metabolism caused by environmental factors (including diet and toxins), disease processes, and the involvement of extragenomic influences, such as gut microflora. This is not a trivial difference; metabolomic studies should, by definition, exclude metabolic contributions from extragenomic sources, because these are external to the system being studied. However, in practice, within the field of human disease research there is still a large degree of overlap in the way both terms are used, and they are often in effect synonymous.[42]
Exometabolomics
Exometabolomics, or "metabolic footprinting", is the study of extracellular metabolites. It uses many techniques from other subfields of metabolomics, and has applications in biofuel development, bioprocessing, determining drugs' mechanism of action, and studying intercellular interactions.[43]
Analytical technologies
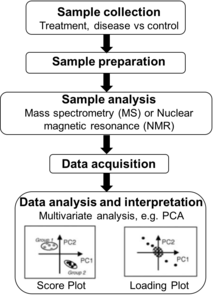
The typical workflow of metabolomics studies is shown in the figure. First, samples are collected from tissue, plasma, urine, saliva, cells, etc. Next, metabolites extracted often with the addition of internal standards and derivatization.[44] During sample analysis, metabolites are quantified (liquid chromatography or gas chromatography coupled with MS and/or NMR spectroscopy). The raw output data can be used for metabolite feature extraction and further processed before statistical analysis (such as PCA). Many bioinformatic tools and software are available to identify associations with disease states and outcomes, determine significant correlations, and characterize metabolic signatures with existing biological knowledge.[45]
Separation methods
Initially, analytes in a metabolomic sample comprise a highly complex mixture. This complex mixture can be simplified prior to detection by separating some analytes from others. Separation achieves various goals: analytes which cannot be resolved by the detector may be separated in this step; in MS analysis ion suppression is reduced; the retention time of the analyte serves as information regarding its identity. This separation step is not mandatory and is often omitted in NMR and "shotgun" based approaches such as shotgun lipidomics.
Gas chromatography (GC), especially when interfaced with mass spectrometry (GC-MS), is a widely used separation technique for metabolomic analysis.[46] GC offers very high chromatographic resolution, and can be used in conjunction with a flame ionization detector (GC/FID) or a mass spectrometer (GC-MS). The method is especially useful for identification and quantification of small and volatile molecules.[47] However, a practical limitation of GC is the requirement of chemical derivatization for many biomolecules as only volatile chemicals can be analysed without derivatization. In cases where greater resolving power is required, two-dimensional chromatography (GCxGC) can be applied.
High performance liquid chromatography (HPLC) has emerged as the most common separation technique for metabolomic analysis. With the advent of electrospray ionization, HPLC was coupled to MS. In contrast with GC, HPLC has lower chromatographic resolution, but requires no derivatization for polar molecules, and separates molecules in the liquid phase. Additionally HPLC has the advantage that a much wider range of analytes can be measured with a higher sensitivity than GC methods.[48]
Capillary electrophoresis (CE) has a higher theoretical separation efficiency than HPLC (although requiring much more time per separation), and is suitable for use with a wider range of metabolite classes than is GC. As for all electrophoretic techniques, it is most appropriate for charged analytes.[49]
Detection methods

Mass spectrometry (MS) is used to identify and to quantify metabolites after optional separation by GC, HPLC (LC-MS), or CE. GC-MS was the first hyphenated technique to be developed. Identification leverages the distinct patterns in which analytes fragment which can be thought of as a mass spectral fingerprint; libraries exist that allow identification of a metabolite according to this fragmentation pattern . MS is both sensitive and can be very specific. There are also a number of techniques which use MS as a stand-alone technology: the sample is infused directly into the mass spectrometer with no prior separation, and the MS provides sufficient selectivity to both separate and to detect metabolites.
For analysis by mass spectrometry the analytes must be imparted with a charge and transferred to the gas phase. Electron ionization (EI) is the most common ionization technique applies to GC separations as it is amenable to low pressures. EI also produces fragmentation of the analyte, both providing structural information while increasing the complexity of the data and possibly obscuring the molecular ion. Atmospheric-pressure chemical ionization (APCI) is an atmospheric pressure technique that can be applied to all the above separation techniques. APCI is a gas phase ionization method slightly more aggressive ionization than ESI which is suitable for less polar compounds. Electrospray ionization (ESI) is the most common ionization technique applied in LC/MS. This soft ionization is most successful for polar molecules with ionizable functional groups. Another commonly used soft ionization technique is secondary electrospray ionization (SESI).
Surface-based mass analysis has seen a resurgence in the past decade, with new MS technologies focused on increasing sensitivity, minimizing background, and reducing sample preparation. The ability to analyze metabolites directly from biofluids and tissues continues to challenge current MS technology, largely because of the limits imposed by the complexity of these samples, which contain thousands to tens of thousands of metabolites. Among the technologies being developed to address this challenge is Nanostructure-Initiator MS (NIMS),[50][51] a desorption/ ionization approach that does not require the application of matrix and thereby facilitates small-molecule (i.e., metabolite) identification. MALDI is also used however, the application of a MALDI matrix can add significant background at <1000 Da that complicates analysis of the low-mass range (i.e., metabolites). In addition, the size of the resulting matrix crystals limits the spatial resolution that can be achieved in tissue imaging. Because of these limitations, several other matrix-free desorption/ionization approaches have been applied to the analysis of biofluids and tissues.
Secondary ion mass spectrometry (SIMS) was one of the first matrix-free desorption/ionization approaches used to analyze metabolites from biological samples. SIMS uses a high-energy primary ion beam to desorb and generate secondary ions from a surface. The primary advantage of SIMS is its high spatial resolution (as small as 50 nm), a powerful characteristic for tissue imaging with MS. However, SIMS has yet to be readily applied to the analysis of biofluids and tissues because of its limited sensitivity at >500 Da and analyte fragmentation generated by the high-energy primary ion beam. Desorption electrospray ionization (DESI) is a matrix-free technique for analyzing biological samples that uses a charged solvent spray to desorb ions from a surface. Advantages of DESI are that no special surface is required and the analysis is performed at ambient pressure with full access to the sample during acquisition. A limitation of DESI is spatial resolution because "focusing" the charged solvent spray is difficult. However, a recent development termed laser ablation ESI (LAESI) is a promising approach to circumvent this limitation. Most recently, ion trap techniques such as orbitrap mass spectrometry are also applied to metabolomics research.[52]
Nuclear magnetic resonance (NMR) spectroscopy is the only detection technique which does not rely on separation of the analytes, and the sample can thus be recovered for further analyses. All kinds of small molecule metabolites can be measured simultaneously - in this sense, NMR is close to being a universal detector. The main advantages of NMR are high analytical reproducibility and simplicity of sample preparation. Practically, however, it is relatively insensitive compared to mass spectrometry-based techniques.[53][54] Comparison of most common used metabolomics methods is shown in the table.
Although NMR and MS are the most widely used, modern day techniques other methods of detection that have been used. These include Fourier-transform ion cyclotron resonance,[55] ion-mobility spectrometry,[56] electrochemical detection (coupled to HPLC), Raman spectroscopy and radiolabel (when combined with thin-layer chromatography).
Statistical methods

The data generated in metabolomics usually consist of measurements performed on subjects under various conditions. These measurements may be digitized spectra, or a list of metabolite features. In its simplest form this generates a matrix with rows corresponding to subjects and columns corresponding with metabolite features (or vice versa).[6] Several statistical programs are currently available for analysis of both NMR and mass spectrometry data. A great number of free software are already available for the analysis of metabolomics data shown in the table. Some statistical tools listed in the table were designed for NMR data analyses were also useful for MS data.[57] For mass spectrometry data, software is available that identifies molecules that vary in subject groups on the basis of mass-over-charge value and sometimes retention time depending on the experimental design.[58]
Once metabolite data matrix is determined, unsupervised data reduction techniques (e.g. PCA) can be used to elucidate patterns and connections. In many studies, including those evaluating drug-toxicity and some disease models, the metabolites of interest are not known a priori. This makes unsupervised methods, those with no prior assumptions of class membership, a popular first choice. The most common of these methods includes principal component analysis (PCA) which can efficiently reduce the dimensions of a dataset to a few which explain the greatest variation.[32] When analyzed in the lower-dimensional PCA space, clustering of samples with similar metabolic fingerprints can be detected. PCA algorithms aim to replace all correlated variables by a much smaller number of uncorrelated variables (referred to as principal components (PCs)) and retain most of the information in the original dataset.[59] This clustering can elucidate patterns and assist in the determination of disease biomarkers - metabolites that correlate most with class membership.
Linear models are commonly used for metabolomics data, but are affected by multicollinearity. On the other hand, multivariate statistics are thriving methods for high-dimensional correlated metabolomics data, of which the most popular one is Projection to Latent Structures (PLS) regression and its classification version PLS-DA. Other data mining methods, such as random forest, support-vector machines, etc. are received increasing attention for untargeted metabolomics data analysis.[60] In the case of univariate methods, variables are analyzed one by one using classical statistics tools (such as Student's t-test, ANOVA or mixed models) and only these with sufficient small p-values are considered relevant.[31] However, correction strategies should be used to reduce false discoveries when multiple comparisons are conducted. For multivariate analysis, models should always be validated to ensure the results can be generalized.
Machine learning is also a powerful tool that can be used in metabolomics analysis. Recently, the authors of a paper published in Analytical Chemistry, developed a retention time prediction software called Retip. This tool, developed in collaboration with NGALAB, the West Coast Metabolomics Centre and Riken empower all labs to apply artificial intelligence to the retention time prediction of small molecules in complex matrix, as human plasma, plants, food or microbials. Retention time prediction increases the identification rate in liquid chromatography and subsequently leads to an improved biological interpretation of metabolomics data[61].
Key applications
Toxicity assessment/toxicology by metabolic profiling (especially of urine or blood plasma samples) detects the physiological changes caused by toxic insult of a chemical (or mixture of chemicals). In many cases, the observed changes can be related to specific syndromes, e.g. a specific lesion in liver or kidney. This is of particular relevance to pharmaceutical companies wanting to test the toxicity of potential drug candidates: if a compound can be eliminated before it reaches clinical trials on the grounds of adverse toxicity, it saves the enormous expense of the trials.[42]
For functional genomics, metabolomics can be an excellent tool for determining the phenotype caused by a genetic manipulation, such as gene deletion or insertion. Sometimes this can be a sufficient goal in itself—for instance, to detect any phenotypic changes in a genetically modified plant intended for human or animal consumption. More exciting is the prospect of predicting the function of unknown genes by comparison with the metabolic perturbations caused by deletion/insertion of known genes. Such advances are most likely to come from model organisms such as Saccharomyces cerevisiae and Arabidopsis thaliana. The Cravatt laboratory at The Scripps Research Institute has recently applied this technology to mammalian systems, identifying the N-acyltaurines as previously uncharacterized endogenous substrates for the enzyme fatty acid amide hydrolase (FAAH) and the monoalkylglycerol ethers (MAGEs) as endogenous substrates for the uncharacterized hydrolase KIAA1363.[62][63]
Metabologenomics is a novel approach to integrate metabolomics and genomics data by correlating microbial-exported metabolites with predicted biosynthetic genes.[64] This bioinformatics-based pairing method enables natural product discovery at a larger-scale by refining non-targeted metabolomic analyses to identify small molecules with related biosynthesis and to focus on those that may not have previously well known structures.
Fluxomics is a further development of metabolomics. The disadvantage of metabolomics is that it only provides the user with steady-state level information, while fluxomics determines the reaction rates of metabolic reactions and can trace metabolites in a biological system over time.
Nutrigenomics is a generalised term which links genomics, transcriptomics, proteomics and metabolomics to human nutrition. In general a metabolome in a given body fluid is influenced by endogenous factors such as age, sex, body composition and genetics as well as underlying pathologies. The large bowel microflora are also a very significant potential confounder of metabolic profiles and could be classified as either an endogenous or exogenous factor. The main exogenous factors are diet and drugs. Diet can then be broken down to nutrients and non-nutrients. Metabolomics is one means to determine a biological endpoint, or metabolic fingerprint, which reflects the balance of all these forces on an individual's metabolism.[65]
See also
References
- Daviss B (April 2005). "Growing pains for metabolomics". The Scientist. 19 (8): 25–28.
- Jordan KW, Nordenstam J, Lauwers GY, Rothenberger DA, Alavi K, Garwood M, Cheng LL (March 2009). "Metabolomic characterization of human rectal adenocarcinoma with intact tissue magnetic resonance spectroscopy". Diseases of the Colon and Rectum. 52 (3): 520–5. doi:10.1007/DCR.0b013e31819c9a2c. PMC 2720561. PMID 19333056.
- Hollywood K, Brison DR, Goodacre R (September 2006). "Metabolomics: current technologies and future trends". Proteomics. 6 (17): 4716–23. doi:10.1002/pmic.200600106. PMID 16888765.
- Gates SC, Sweeley CC (October 1978). "Quantitative metabolic profiling based on gas chromatography". Clinical Chemistry. 24 (10): 1663–73. doi:10.1093/clinchem/24.10.1663. PMID 359193.
- Preti, George (June 6, 2005). "Metabolomics comes of age?". The Scientist. 19 (11): 8.
- Van der greef and Smilde, J Chemomet, (2005) 19:376-386
- Shapiro I, Kavkalo DN, Petrova GV, Ganzin AP (2008). "[Angioleiomyoma of the large-intestinal mesentery complicated by diffuse peritonitis]". Sovetskaia Meditsina. 866 (9): 26–47. doi:10.1016/j.jchromb.2007.10.007. PMC 2603028. PMID 18551752.
- Griffiths WJ, Wang Y (July 2009). "Mass spectrometry: from proteomics to metabolomics and lipidomics". Chemical Society Reviews. 38 (7): 1882–96. doi:10.1039/b618553n. PMID 19551169.
- Hoult DI, Busby SJ, Gadian DG, Radda GK, Richards RE, Seeley PJ (November 1974). "Observation of tissue metabolites using 31P nuclear magnetic resonance". Nature. 252 (5481): 285–7. Bibcode:1974Natur.252..285H. doi:10.1038/252285a0. PMID 4431445.
- Nicholson JK, Lindon JC (October 2008). "Systems biology: Metabonomics". Nature. 455 (7216): 1054–6. Bibcode:2008Natur.455.1054N. doi:10.1038/4551054a. PMID 18948945.
- Holmes E, Antti H (December 2002). "Chemometric contributions to the evolution of metabonomics: mathematical solutions to characterising and interpreting complex biological NMR spectra". The Analyst. 127 (12): 1549–57. Bibcode:2002Ana...127.1549H. doi:10.1039/b208254n. PMID 12537357.
- Lenz EM, Wilson ID (February 2007). "Analytical strategies in metabonomics". Journal of Proteome Research. 6 (2): 443–58. doi:10.1021/pr0605217. PMID 17269702.
- Cravatt BF, Prospero-Garcia O, Siuzdak G, Gilula NB, Henriksen SJ, Boger DL, Lerner RA (June 1995). "Chemical characterization of a family of brain lipids that induce sleep". Science. 268 (5216): 1506–9. Bibcode:1995Sci...268.1506C. doi:10.1126/science.7770779. PMID 7770779.
- Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, et al. (December 2005). "METLIN: a metabolite mass spectral database". Therapeutic Drug Monitoring. 27 (6): 747–51. doi:10.1097/01.ftd.0000179845.53213.39. PMID 16404815.
- Guijas C, Montenegro-Burke JR, Domingo-Almenara X, Palermo A, Warth B, Hermann G, et al. (March 2018). "METLIN: A Technology Platform for Identifying Knowns and Unknowns". Analytical Chemistry. 90 (5): 3156–3164. doi:10.1021/acs.analchem.7b04424. PMC 5933435. PMID 29381867.
- Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G (February 2006). "XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification". Analytical Chemistry. 78 (3): 779–87. doi:10.1021/ac051437y. PMID 16448051.
- Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G (June 2012). "XCMS Online: a web-based platform to process untargeted metabolomic data". Analytical Chemistry. 84 (11): 5035–9. doi:10.1021/ac300698c. PMC 3703953. PMID 22533540.
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. (January 2007). "HMDB: the Human Metabolome Database". Nucleic Acids Research. 35 (Database issue): D521-6. doi:10.1093/nar/gkl923. PMC 1899095. PMID 17202168.
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, et al. (January 2009). "HMDB: a knowledgebase for the human metabolome". Nucleic Acids Research. 37 (Database issue): D603-10. doi:10.1093/nar/gkn810. PMC 2686599. PMID 18953024.
- Farag MA, Huhman DV, Dixon RA, Sumner LW (February 2008). "Metabolomics reveals novel pathways and differential mechanistic and elicitor-specific responses in phenylpropanoid and isoflavonoid biosynthesis in Medicago truncatula cell cultures". Plant Physiology. 146 (2): 387–402. doi:10.1104/pp.107.108431. PMC 2245840. PMID 18055588.
- "www.Plantmetabolomics.org". Nov 7, 2012. Archived from the original on 2012-11-07. Retrieved May 20, 2020.
- Morrow Jr KJ (1 April 2010). "Mass Spec Central to Metabolomics". Genetic Engineering & Biotechnology News. 30 (7). p. 1. Archived from the original on 28 June 2010. Retrieved 28 June 2010.
- "Real-time analysis of metabolic products". phys.org. Retrieved May 20, 2020.
- Querengesser, Lori; Vogel, Hans J.; Sykes, Brian D.; Marrie, Tom; Li, Liang; Greiner, Russ; Clive, Derrick; Bamforth, Fiona; Dowlatabadi, Reza (2007-01-01). "HMDB: the Human Metabolome Database". Nucleic Acids Research. 35 (suppl_1): D521–D526. doi:10.1093/nar/gkl923. ISSN 0305-1048. PMC 1899095. PMID 17202168.
- Oliver SG, Winson MK, Kell DB, Baganz F (September 1998). "Systematic functional analysis of the yeast genome". Trends in Biotechnology. 16 (9): 373–8. doi:10.1016/S0167-7799(98)01214-1. PMID 9744112.
- Griffin JL, Vidal-Puig A (June 2008). "Current challenges in metabolomics for diabetes research: a vital functional genomic tool or just a ploy for gaining funding?". Physiological Genomics. 34 (1): 1–5. doi:10.1152/physiolgenomics.00009.2008. PMID 18413782.
- HMDB 3.0 – the human metabolome database in 2013.
- Pearson H (March 2007). "Meet the human metabolome". Nature. 446 (7131): 8. Bibcode:2007Natur.446....8P. doi:10.1038/446008a. PMID 17330009.
- De Luca V, St Pierre B (April 2000). "The cell and developmental biology of alkaloid biosynthesis". Trends in Plant Science. 5 (4): 168–73. doi:10.1016/S1360-1385(00)01575-2. PMID 10740298.
- Griffin JL, Shockcor JP (July 2004). "Metabolic profiles of cancer cells". Nature Reviews. Cancer. 4 (7): 551–61. doi:10.1038/nrc1390. PMID 15229480.
- Ulaszewska, Marynka M.; Weinert, Christoph H.; Trimigno, Alessia; Portmann, Reto; Andres Lacueva, Cristina; Badertscher, René; Brennan, Lorraine; Brunius, Carl; Bub, Achim; Capozzi, Francesco; Cialiè Rosso, Marta; Cordero, Chiara E.; Daniel, Hannelore; Durand, Stéphanie; Egert, Bjoern; Ferrario, Paola G.; Feskens, Edith J.M.; Franceschi, Pietro; Garcia-Aloy, Mar; Giacomoni, Franck; Giesbertz, Pieter; González-Domínguez, Raúl; Hanhineva, Kati; Hemeryck, Lieselot Y.; Kopka, Joachim; Kulling, Sabine E.; Llorach, Rafael; Manach, Claudine; Mattivi, Fulvio; et al. (2019). "Nutrimetabolomics: An Integrative Action for Metabolomic Analyses in Human Nutritional Studies". Molecular Nutrition and Food Research. 63 (1): 1800384. doi:10.1002/mnfr.201800384. PMID 30176196.
- Nicholson JK, Wilson ID (August 2003). "Opinion: understanding 'global' systems biology: metabonomics and the continuum of metabolism". Nature Reviews. Drug Discovery. 2 (8): 668–76. doi:10.1038/nrd1157. PMID 12904817.
- Dettmer, Katja; Aronov, Pavel A.; Hammock, Bruce D. (2007). "Mass spectrometry-based metabolomics". Mass Spectrometry Reviews. 26 (1): 51–78. Bibcode:2007MSRv...26...51D. doi:10.1002/mas.20108. ISSN 0277-7037. PMC 1904337. PMID 16921475.
- Nicholson JK, Foxall PJ, Spraul M, Farrant RD, Lindon JC (March 1995). "750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma". Analytical Chemistry. 67 (5): 793–811. doi:10.1021/ac00101a004. PMID 7762816.
- Bentley R (1999). "Secondary metabolite biosynthesis: the first century". Critical Reviews in Biotechnology. 19 (1): 1–40. doi:10.1080/0738-859991229189. PMID 10230052.
- Nordström A, O'Maille G, Qin C, Siuzdak G (May 2006). "Nonlinear data alignment for UPLC-MS and HPLC-MS based metabolomics: quantitative analysis of endogenous and exogenous metabolites in human serum". Analytical Chemistry. 78 (10): 3289–95. doi:10.1021/ac060245f. PMC 3705959. PMID 16689529.
- Crockford DJ, Maher AD, Ahmadi KR, Barrett A, Plumb RS, Wilson ID, Nicholson JK (September 2008). "1H NMR and UPLC-MS(E) statistical heterospectroscopy: characterization of drug metabolites (xenometabolome) in epidemiological studies". Analytical Chemistry. 80 (18): 6835–44. doi:10.1021/ac801075m. PMID 18700783.
- Nicholson JK (2006). "Global systems biology, personalized medicine and molecular epidemiology". Molecular Systems Biology. 2 (1): 52. doi:10.1038/msb4100095. PMC 1682018. PMID 17016518.
- Nicholson JK, Lindon JC, Holmes E (November 1999). "'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data". Xenobiotica; the Fate of Foreign Compounds in Biological Systems. 29 (11): 1181–9. doi:10.1080/004982599238047. PMID 10598751.
- Nicholson JK, Connelly J, Lindon JC, Holmes E (February 2002). "Metabonomics: a platform for studying drug toxicity and gene function". Nature Reviews. Drug Discovery. 1 (2): 153–61. doi:10.1038/nrd728. PMID 12120097.
- Holmes E, Wilson ID, Nicholson JK (September 2008). "Metabolic phenotyping in health and disease". Cell. 134 (5): 714–7. doi:10.1016/j.cell.2008.08.026. PMID 18775301.
- Robertson DG (June 2005). "Metabonomics in toxicology: a review". Toxicological Sciences. 85 (2): 809–22. doi:10.1093/toxsci/kfi102. PMID 15689416.
- Silva LP, Northen TR (August 2015). "Exometabolomics and MSI: deconstructing how cells interact to transform their small molecule environment". Current Opinion in Biotechnology. Elsevier BV. 34: 209–16. doi:10.1016/j.copbio.2015.03.015. PMID 25855407.
- Dettmer K, Aronov PA, Hammock BD (2007). "Mass spectrometry-based metabolomics". Mass Spectrometry Reviews. 26 (1): 51–78. Bibcode:2007MSRv...26...51D. doi:10.1002/mas.20108. PMC 1904337. PMID 16921475.
- Rasmiena AA, Ng TW, Meikle PJ (March 2013). "Metabolomics and ischaemic heart disease". Clinical Science. 124 (5): 289–306. doi:10.1042/CS20120268. PMID 23157406.
- Ogbaga CC, Stepien P, Dyson BC, Rattray NJ, Ellis DI, Goodacre R, Johnson GN (6 May 2016). "Biochemical Analyses of Sorghum Varieties Reveal Differential Responses to Drought". PLOS ONE. 11 (5): e0154423. Bibcode:2016PLoSO..1154423O. doi:10.1371/journal.pone.0154423. PMC 4859509. PMID 27153323.
- "Gas Chromatography Mass Spectrometry (GC-MS) Information | Thermo Fisher Scientific - US". www.thermofisher.com. Retrieved 2018-09-26.
- Gika HG, Theodoridis GA, Wingate JE, Wilson ID (August 2007). "Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: application to human urine". Journal of Proteome Research. 6 (8): 3291–303. doi:10.1021/pr070183p. PMID 17625818.
- Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T (September 2003). "Quantitative metabolome analysis using capillary electrophoresis mass spectrometry". Journal of Proteome Research. 2 (5): 488–94. doi:10.1021/pr034020m. PMID 14582645.
- Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, et al. (October 2007). "Clathrate nanostructures for mass spectrometry". Nature. 449 (7165): 1033–6. Bibcode:2007Natur.449.1033N. doi:10.1038/nature06195. PMID 17960240.
- Woo HK, Northen TR, Yanes O, Siuzdak G (July 2008). "Nanostructure-initiator mass spectrometry: a protocol for preparing and applying NIMS surfaces for high-sensitivity mass analysis". Nature Protocols. 3 (8): 1341–9. doi:10.1038/NPROT.2008.110. PMID 18714302.
- Ghaste, Manoj; Mistrik, Robert; Shulaev, Vladimir (June 2016). "Applications of Fourier Transform Ion Cyclotron Resonance (FT-ICR) and Orbitrap Based High Resolution Mass Spectrometry in Metabolomics and Lipidomics". International Journal of Molecular Sciences. 17 (6): 816. doi:10.3390/ijms17060816. PMC 4926350. PMID 27231903.
- Griffin JL (October 2003). "Metabonomics: NMR spectroscopy and pattern recognition analysis of body fluids and tissues for characterisation of xenobiotic toxicity and disease diagnosis". Current Opinion in Chemical Biology. 7 (5): 648–54. doi:10.1016/j.cbpa.2003.08.008. PMID 14580571.
- Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, Nicholson JK (2007). "Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts". Nature Protocols. 2 (11): 2692–703. doi:10.1038/nprot.2007.376. PMID 18007604.
- Habchi, Baninia; Alves, Sandra; Jouan-Rimbaud Bouveresse, Delphine; Appenzeller, Brice; Paris, Alain; Rutledge, Douglas N.; Rathahao-Paris, Estelle (2018-01-01). "Potential of dynamically harmonized Fourier transform ion cyclotron resonance cell for high-throughput metabolomics fingerprinting: control of data quality". Analytical and Bioanalytical Chemistry. 410 (2): 483–490. doi:10.1007/s00216-017-0738-3. ISSN 1618-2650. PMID 29167936.
- King, Adam M.; Mullin, Lauren G.; Wilson, Ian D.; Coen, Muireann; Rainville, Paul D.; Plumb, Robert S.; Gethings, Lee A.; Maker, Garth; Trengove, Robert (2019-01-22). "Development of a rapid profiling method for the analysis of polar analytes in urine using HILIC–MS and ion mobility enabled HILIC–MS". Metabolomics. 15 (2): 17. doi:10.1007/s11306-019-1474-9. ISSN 1573-3890. PMC 6342856. PMID 30830424.
- Sugimoto M, Kawakami M, Robert M, Soga T, Tomita M (March 2012). "Bioinformatics Tools for Mass Spectroscopy-Based Metabolomic Data Processing and Analysis". Current Bioinformatics. 7 (1): 96–108. doi:10.2174/157489312799304431. PMC 3299976. PMID 22438836.
- Spicer R, Salek RM, Moreno P, Cañueto D, Steinbeck C (2017). "Navigating freely-available software tools for metabolomics analysis". Metabolomics. 13 (9): 106. doi:10.1007/s11306-017-1242-7. PMC 5550549. PMID 28890673.
- Ren S, Hinzman AA, Kang EL, Szczesniak RD, Lu LJ (December 2015). "Computational and statistical analysis of metabolomics data". Metabolomics. 11 (6): 1492–513. doi:10.1007/s11306-015-0823-6.
- Gromski PS, Muhamadali H, Ellis DI, Xu Y, Correa E, Turner ML, Goodacre R (2015). "A tutorial review: Metabolomics and partial least squares-discriminant analysis – a marriage of convenience or a shotgun wedding". Analytica Chimica Acta. 879: 10–23. doi:10.1016/j.aca.2015.02.012. PMID 26002472.
- Bonini, Paolo; Kind, Tobias; Tsugawa, Hiroshi; Barupal, Dinesh Kumar; Fiehn, Oliver (2020-05-10). "Retip: retention time prediction for compound annotation in untargeted metabolomics". Analytical Chemistry. doi:10.1021/acs.analchem.9b05765. ISSN 0003-2700. PMID 32390414.
- Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF (November 2004). "Assignment of endogenous substrates to enzymes by global metabolite profiling" (PDF). Biochemistry. 43 (45): 14332–9. doi:10.1021/bi0480335. PMID 15533037.
- Chiang KP, Niessen S, Saghatelian A, Cravatt BF (October 2006). "An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling". Chemistry & Biology. 13 (10): 1041–50. doi:10.1016/j.chembiol.2006.08.008. PMID 17052608.
- Goering AW, McClure RA, Doroghazi JR, Albright JC, Haverland NA, Zhang Y, et al. (February 2016). "Metabologenomics: Correlation of Microbial Gene Clusters with Metabolites Drives Discovery of a Nonribosomal Peptide with an Unusual Amino Acid Monomer". ACS Central Science. 2 (2): 99–108. doi:10.1021/acscentsci.5b00331. PMC 4827660. PMID 27163034.
- Gibney MJ, Walsh M, Brennan L, Roche HM, German B, van Ommen B (September 2005). "Metabolomics in human nutrition: opportunities and challenges". The American Journal of Clinical Nutrition. 82 (3): 497–503. doi:10.1093/ajcn/82.3.497. PMID 16155259.
Further reading
- Tomita M, Nishioka T (2005). Metabolomics: The Frontier of Systems Biology. Springer. ISBN 4-431-25121-9.
- Weckwerth W (2006). Metabolomics: Methods And Protocols (Methods in Molecular Biology). Humana Press. ISBN 1-588-29561-3. OCLC 493824826.
- Fan TW, Lorkiewicz PK, Sellers K, Moseley HN, Higashi RM, Lane AN (March 2012). "Stable isotope-resolved metabolomics and applications for drug development". Pharmacology & Therapeutics. 133 (3): 366–91. doi:10.1016/j.pharmthera.2011.12.007. PMC 3471671. PMID 22212615.
- Ellis DI, Goodacre R (August 2006). "Metabolic fingerprinting in disease diagnosis: biomedical applications of infrared and Raman spectroscopy" (PDF). The Analyst. 131 (8): 875–85. Bibcode:2006Ana...131..875E. doi:10.1039/b602376m. PMID 17028718.
- Claudino WM, Quattrone A, Biganzoli L, Pestrin M, Bertini I, Di Leo A (July 2007). "Metabolomics: available results, current research projects in breast cancer, and future applications". Journal of Clinical Oncology. 25 (19): 2840–6. doi:10.1200/JCO.2006.09.7550. PMID 17502626. Archived from the original on 2008-01-20.
- Ellis DI, Dunn WB, Griffin JL, Allwood JW, Goodacre R (September 2007). "Metabolic fingerprinting as a diagnostic tool" (PDF). Pharmacogenomics. 8 (9): 1243–66. doi:10.2217/14622416.8.9.1243. PMID 17924839.
- Bundy, JG; Davey, MP; Viant, MR (2009). "Environmental metabolomics: A critical review and future perspectives". Metabolomics. 5: 3–21. doi:10.1007/s11306-008-0152-0.
- Haug K, Salek RM, Conesa P, Hastings J, de Matos P, Rijnbeek M, et al. (January 2013). "MetaboLights--an open-access general-purpose repository for metabolomics studies and associated meta-data". Nucleic Acids Research. 41 (Database issue): D781-6. doi:10.1093/nar/gks1004. PMC 3531110. PMID 23109552.
External links
| Look up metabolomics in Wiktionary, the free dictionary. |
| Wikibooks has more on the topic of: Metabolomics |