Measles morbillivirus
Measles morbillivirus, formerly called measles virus (MV), is a single-stranded, negative-sense, enveloped, non-segmented RNA virus of the genus Morbillivirus within the family Paramyxoviridae. It is the cause of measles. Humans are the natural hosts of the virus; no animal reservoirs are known to exist.
| Measles morbillivirus | |
|---|---|
| Measles morbillivirus electron micrograph | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Monjiviricetes |
| Order: | Mononegavirales |
| Family: | Paramyxoviridae |
| Genus: | Morbillivirus |
| Species: | Measles morbillivirus |
| Synonyms[1] | |
|
Measles virus | |
Disease
The virus causes measles, an infection of the respiratory system. Symptoms include fever, cough, runny nose, red eyes and a generalized, maculopapular, erythematous rash. The virus is highly contagious and is spread by coughing and sneezing via close personal contact or direct contact with secretions.
Replication cycle
Entry
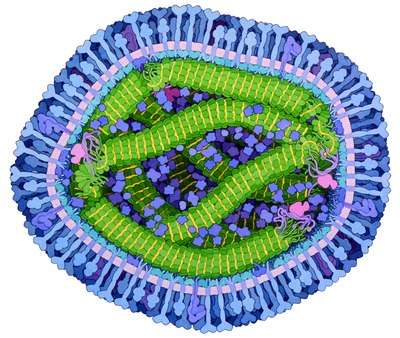
The measles virus has two envelope glycoproteins on the viral surface – hemagglutinin (H) and membrane fusion protein (F). These proteins are responsible for host cell binding and invasion. Three receptors for the H protein have been identified to date: complement regulatory molecule CD46, the signaling lymphocyte activation molecule (SLAM) and the cell adhesion molecule Nectin-4.[2]
Genome replication and viral assembly
Once the virus has entered a host cell, its strand of negative sense ssRNA (single stranded RNA) is used as a template to create a positive sense copy, using the RNA-dependent RNA polymerase that's included in the virion. Then this copy is used to create a new negative copy, and so on, to create many copies of the ssRNA. The positive sense ssRNA is then mass translated by host ribosomes, producing all viral proteins. The viruses are then assembled from their proteins and negative sense ssRNA, and the cell will lyse, discharging the new viral particles and restarting the cycle.[3]
Evolution
The measles virus evolved from the now eradicated but formerly widespread rinderpest virus, which infected cattle.[4] Sequence analysis has suggested that the two viruses most probably diverged in the 11th and 12th centuries, though the periods as early as the 5th century fall within the 95% confidence interval of these calculations.[4]
Other analysis has suggested that the divergence may be even older because of the technique's tendency to underestimate ages when strong purifying selection is in action.[5] There is some linguistic evidence for an earlier origin within the seventh century.[6][7] The current epidemic strain evolved at the beginning of the 20th century—most probably between 1908 and 1943.[8]
Genotypes
The measles virus genome is typically 15,894 nucleotides long and encodes eight proteins.[9] The WHO currently recognises 8 clades of measles (A–H). Subtypes are designed with numerals—A1, D2 etc. Currently, 23 subtypes are recognised. The 450 nucleotides that code for the C‐terminal 150 amino acids of N are the minimum amount of sequence data required for genotyping a measles virus isolate. The genotyping scheme was introduced in 1998 and extended in 2002 and 2003.
Despite the variety of measles genotypes, there is only one measles serotype. Antibodies to measles bind to the hemagglutinin protein. Thus, antibodies against one genotype (such as the vaccine strain) protect against all other genotypes.[10]
The major genotypes differ between countries and the status of measles circulation within that country or region. Endemic transmission of measles virus was interrupted in the United States and Australia by 2000 and the Americas by 2002.[11]
References
- "ICTV Taxonomy history: Measles morbillivirus" (html). International Committee on Taxonomy of Viruses (ICTV). Retrieved 15 January 2019.
- Lu G, Gao GF, Yan J (2013). "The receptors and entry of measles virus: a review". Sheng Wu Gong Cheng Xue Bao (in Chinese). 29 (1): 1–9. PMID 23631113.
- Jiang, Yanliang; Qin, Yali; Chen, Mingzhou (16 November 2016). "Host–Pathogen Interactions in Measles Virus Replication and Anti-Viral Immunity". Viruses. 8 (11): 308. doi:10.3390/v8110308. ISSN 1999-4915. PMC 5127022. PMID 27854326.
- Furuse Y, Suzuki A, Oshitani H (2010). "Origin of measles virus: divergence from rinderpest virus between the 11th and 12th centuries". Virol. J. 7: 52. doi:10.1186/1743-422X-7-52. PMC 2838858. PMID 20202190.
- Wertheim, J. O.; Kosakovsky Pond, S. L. (2011). "Purifying Selection Can Obscure the Ancient Age of Viral Lineages". Molecular Biology and Evolution. 28 (12): 3355–65. doi:10.1093/molbev/msr170. PMC 3247791. PMID 21705379.
- Griffin DE (2007). "Measles Virus". In Martin, Malcolm A.; Knipe, David M.; Fields, Bernard N.; Howley, Peter M.; Griffin, Diane; Lamb, Robert (eds.). Fields' virology (5th ed.). Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins. ISBN 978-0-7817-6060-7.
- McNeil, W. (1976). Plagues and Peoples. New York: Anchor Press/Doubleday. ISBN 978-0-385-11256-7.
- Pomeroy LW, Bjørnstad ON, Holmes EC (February 2008). "The evolutionary and epidemiological dynamics of the paramyxoviridae". J. Mol. Evol. 66 (2): 98–106. Bibcode:2008JMolE..66...98P. doi:10.1007/s00239-007-9040-x. PMC 3334863. PMID 18217182.
- Phan, MVT; Schapendonk, CME; Oude Munnink, BB; Koopmans, MPG; de Swart, RL; Cotten, M (29 March 2018). "Complete Genome Sequences of Six Measles Virus Strains". Genome Announcements. 6 (13). doi:10.1128/genomeA.00184-18. PMC 5876482. PMID 29599155.
- "Measles". Biologicals — Vaccine Standardization. World Health Organization. 11 March 2013.
- "No endemic transmission of measles in the Americas since 2002". Pan American Health Organization. 14 February 2017. Retrieved 11 September 2017.
External links
| Wikimedia Commons has media related to Measles virus. |
| Wikispecies has information related to Measles morbillivirus |
- "Measles virus". International Committee on Taxonomy of Viruses.