Hydrogen bond
A hydrogen bond (often informally abbreviated H-bond) is a partial intermolecular bonding interaction between a lone pair on an electron rich donor atom, particularly the second-row elements nitrogen (N), oxygen (O), or fluorine (F), and the antibonding molecular orbital of a bond between hydrogen (H) and a more electronegative atom or group.[4] Such an interacting system is generally denoted Dn–H···Ac, where the solid line denotes a polar covalent bond, and the dotted or dashed line indicates the hydrogen bond. The use of three centered dots for the hydrogen bond is specifically recommended by the IUPAC.[5] While hydrogen bonding has both covalent and electrostatic contributions, and the degrees to which they contribute are currently debated, the present evidence strongly implies that the primary contribution is covalent.[6]
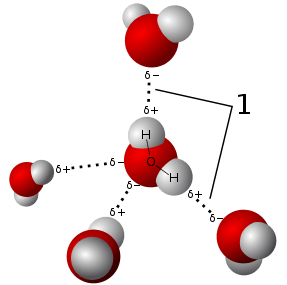
Hydrogen bonds can be intermolecular (occurring between separate molecules) or intramolecular (occurring among parts of the same molecule).[7][8][9][10] Depending on the nature of the donor and acceptor atoms which constitute the bond, their geometry, and environment, the energy of a hydrogen bond can vary between 1 and 40 kcal/mol.[11] This makes them somewhat stronger than a van der Waals interaction, and weaker than fully covalent or ionic bonds. This type of bond can occur in inorganic molecules such as water and in organic molecules like DNA and proteins.
The hydrogen bond is responsible for many of the anomalous physical and chemical properties of compounds of N, O, and F. In particular, intermolecular hydrogen bonding is responsible for the high boiling point of water (100 °C) compared to the other group 16 hydrides that have much weaker hydrogen bonds.[12] Intramolecular hydrogen bonding is partly responsible for the secondary and tertiary structures of proteins and nucleic acids. It also plays an important role in the structure of polymers, both synthetic and natural.
Weaker hydrogen bonds[13] are known for hydrogen atoms bound to elements such as sulfur (S) or chlorine (Cl); even carbon (C) can serve as a donor, particularly when the carbon or one of its neighbors is electronegative (e.g., in chloroform, aldehydes and terminal acetylenes).[14][15] Gradually, it was recognized that there are many examples of weaker hydrogen bonding involving donor other than N, O, or F and/or acceptor Ac with electronegativity approaching that of hydrogen (rather than being much more electronegative). Though these "non-traditional" hydrogen bonding interactions are often quite weak (~1 kcal/mol), they are also ubiquitous and are increasingly recognized as important control elements in receptor-ligand interactions in medicinal chemistry or intra-/intermolecular interactions in materials sciences. The definition of hydrogen bonding has gradually broadened over time to include these weaker attractive interactions. In 2011, an IUPAC Task Group recommended a modern evidence-based definition of hydrogen bonding, which was published in the IUPAC journal Pure and Applied Chemistry. This definition specifies:
The hydrogen bond is an attractive interaction between a hydrogen atom from a molecule or a molecular fragment X–H in which X is more electronegative than H, and an atom or a group of atoms in the same or a different molecule, in which there is evidence of bond formation.[16]
As part of a more detailed list of criteria, the IUPAC publication acknowledges that the attractive interaction can arise from some combination of electrostatics (multipole-multipole and multipole-induced multipole interactions), covalency (charge transfer by orbital overlap), and dispersion (London forces), and states that the relative importance of each will vary depending on the system. However, a footnote to the criterion recommends the exclusion of interactions in which dispersion is the primary contributor, specifically giving Ar---CH4 and CH4---CH4 as examples of such interactions to be excluded from the definition.[5]
Nevertheless, most introductory textbooks still restrict the definition of hydrogen bond to the "classical" type of hydrogen bond characterized in the opening paragraph.
Bonding
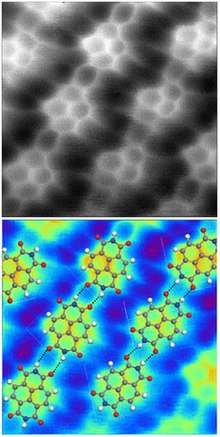
Definitions and general characteristics
A hydrogen atom attached to a relatively electronegative atom is the hydrogen bond donor.[18] C-H bonds only participate in hydrogen bonding when the carbon atom is bound to electronegative substituents, as is the case in chloroform, CHCl3.[19] In a hydrogen bond, the electronegative atom not covalently attached to the hydrogen is named proton acceptor, whereas the one covalently bound to the hydrogen is named the proton donor. While this nomenclature is recommended by the IUPAC[5], it can be misleading, since in other donor-acceptor bonds, the donor/acceptor assignment is based on the source of the electron pair (such nomenclature is also used for hydrogen bonds by some authors[20]). In the hydrogen bond donor, the H center is protic. The donor is a Lewis acid. Hydrogen bonds are represented as H···Y system, where the dots represent the hydrogen bond. Liquids that display hydrogen bonding (such as water) are called associated liquids.
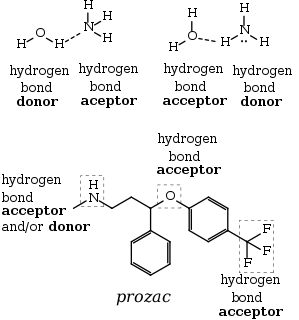
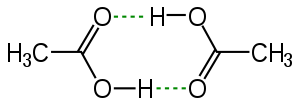
The hydrogen bond is often described as an electrostatic dipole-dipole interaction. However, it also has some features of covalent bonding: it is directional and strong, produces interatomic distances shorter than the sum of the van der Waals radii, and usually involves a limited number of interaction partners, which can be interpreted as a type of valence. These covalent features are more substantial when acceptors bind hydrogens from more electronegative donors.
Bond strength
Hydrogen bonds can vary in strength from weak (1–2 kJ mol−1) to strong (161.5 kJ mol−1 in the ion HF−
2).[21][22] Typical enthalpies in vapor include:[23]
- F−H···:F (161.5 kJ/mol or 38.6 kcal/mol), illustrated uniquely by HF2−, bifluoride
- O−H···:N (29 kJ/mol or 6.9 kcal/mol), illustrated water-ammonia
- O−H···:O (21 kJ/mol or 5.0 kcal/mol), illustrated water-water, alcohol-alcohol
- N−H···:N (13 kJ/mol or 3.1 kcal/mol), illustrated by ammonia-ammonia
- N−H···:O (8 kJ/mol or 1.9 kcal/mol), illustrated water-amide
- HO−H···:OH+
3 (18 kJ/mol[24] or 4.3 kcal/mol)
The strength of intermolecular hydrogen bonds is most often evaluated by measurements of equilibria between molecules containing donor and/or acceptor units, most often in solution.[25] The strength of intramolecular hydrogen bonds can be studied with equilibria between conformers with and without hydrogen bonds. The most important method for the identification of hydrogen bonds also in complicated molecules is crystallography, sometimes also NMR-spectroscopy. Structural details, in particular distances between donor and acceptor which are smaller than the sum of the van der Waals radii can be taken as indication of the hydrogen bond strength.
One scheme gives the following somewhat arbitrary classification: those that are 15 to 40 kcal/mol, 5 to 15 kcal/mol, and >0 to 5 kcal/mol are considered strong, moderate, and weak, respectively.
Structural details
The X−H distance is typically ≈110 pm, whereas the H···Y distance is ≈160 to 200 pm. The typical length of a hydrogen bond in water is 197 pm. The ideal bond angle depends on the nature of the hydrogen bond donor. The following hydrogen bond angles between a hydrofluoric acid donor and various acceptors have been determined experimentally:[26]
| Acceptor···donor | VSEPR geometry | Angle (°) |
|---|---|---|
| HCN···HF | linear | 180 |
| H2CO···HF | trigonal planar | 120 |
| H2O···HF | pyramidal | 46 |
| H2S···HF | pyramidal | 89 |
| SO2···HF | trigonal | 142 |
Spectroscopy
Strong hydrogen bonds are revealed by downfield shifts in the 1H NMR spectrum. For example, the acidic proton in the enol tautomer of acetylacetone appears at δH 15.5, which is about 10 ppm downfield of a conventional alcohol.[27]
In the IR spectrum, hydrogen bonding shifts the X-H stretching frequency to lower energy (i.e. the vibration frequency decreases). This shift reflects a weakening of the X-H bond. Certain hydrogen bonds - improper hydrogen bonds - show a blue shift of the X-H stretching frequency and a decrease in the bond length.[28] H-bonds can also be measured by IR vibrational mode shifts of the acceptor. The amide I mode of backbone carbonyls in α-helices shifts to lower frequencies when they form H-bonds with side-chain hydroxyl groups.[29]
Theoretical considerations
Hydrogen bonding is of continuing theoretical interest. According to a modern description O:H-O integrates both the intermolecular O:H lone pair ":" nonbond and the intramolecular H-O polar-covalent bond associated with O-O repulsive coupling.[30]
Quantum chemical calculations of the relevant interresidue potential constants (compliance constants) revealed large differences between individual H bonds of the same type. For example, the central interresidue N−H···N hydrogen bond between guanine and cytosine is much stronger in comparison to the N−H···N bond between the adenine-thymine pair.[31]
Theoretically, the bond strength of the hydrogen bonds can be assessed using NCI index, non-covalent interactions index, which allows a visualization of these non-covalent interactions, as its name indicates, using the electron density of the system.
From interpretations of the anisotropies in the Compton profile of ordinary ice that the hydrogen bond is partly covalent.[32] However, this interpretation was challenged.[33]
Most generally, the hydrogen bond can be viewed as a metric-dependent electrostatic scalar field between two or more intermolecular bonds. This is slightly different from the intramolecular bound states of, for example, covalent or ionic bonds; however, hydrogen bonding is generally still a bound state phenomenon, since the interaction energy has a net negative sum. The initial theory of hydrogen bonding proposed by Linus Pauling suggested that the hydrogen bonds had a partial covalent nature. This interpretation remained controversial until NMR techniques demonstrated information transfer between hydrogen-bonded nuclei, a feat that would only be possible if the hydrogen bond contained some covalent character.[34]
History
The concept of hydrogen bonding once was challenging.[35] Linus Pauling credits T. S. Moore and T. F. Winmill with the first mention of the hydrogen bond, in 1912.[36][37] Moore and Winmill used the hydrogen bond to account for the fact that trimethylammonium hydroxide is a weaker base than tetramethylammonium hydroxide. The description of hydrogen bonding in its better-known setting, water, came some years later, in 1920, from Latimer and Rodebush.[38] In that paper, Latimer and Rodebush cite work by a fellow scientist at their laboratory, Maurice Loyal Huggins, saying, "Mr. Huggins of this laboratory in some work as yet unpublished, has used the idea of a hydrogen kernel held between two atoms as a theory in regard to certain organic compounds."
Hydrogen bonds in small molecules
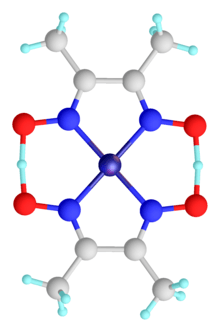
Water
A ubiquitous example of a hydrogen bond is found between water molecules. In a discrete water molecule, there are two hydrogen atoms and one oxygen atom. Two molecules of water can form a hydrogen bond between them that is to say oxygen-hydrogen bonding; the simplest case, when only two molecules are present, is called the water dimer and is often used as a model system. When more molecules are present, as is the case with liquid water, more bonds are possible because the oxygen of one water molecule has two lone pairs of electrons, each of which can form a hydrogen bond with a hydrogen on another water molecule. This can repeat such that every water molecule is H-bonded with up to four other molecules, as shown in the figure (two through its two lone pairs, and two through its two hydrogen atoms). Hydrogen bonding strongly affects the crystal structure of ice, helping to create an open hexagonal lattice. The density of ice is less than the density of water at the same temperature; thus, the solid phase of water floats on the liquid, unlike most other substances.
Liquid water's high boiling point is due to the high number of hydrogen bonds each molecule can form, relative to its low molecular mass. Owing to the difficulty of breaking these bonds, water has a very high boiling point, melting point, and viscosity compared to otherwise similar liquids not conjoined by hydrogen bonds. Water is unique because its oxygen atom has two lone pairs and two hydrogen atoms, meaning that the total number of bonds of a water molecule is up to four.
The number of hydrogen bonds formed by a molecule of liquid water fluctuates with time and temperature.[39] From TIP4P liquid water simulations at 25 °C, it was estimated that each water molecule participates in an average of 3.59 hydrogen bonds. At 100 °C, this number decreases to 3.24 due to the increased molecular motion and decreased density, while at 0 °C, the average number of hydrogen bonds increases to 3.69.[39] A more recent study found a much smaller number of hydrogen bonds: 2.357 at 25 °C.[40] The differences may be due to the use of a different method for defining and counting the hydrogen bonds.
Where the bond strengths are more equivalent, one might instead find the atoms of two interacting water molecules partitioned into two polyatomic ions of opposite charge, specifically hydroxide (OH−) and hydronium (H3O+). (Hydronium ions are also known as "hydroxonium" ions.)
- H−O− H3O+
Indeed, in pure water under conditions of standard temperature and pressure, this latter formulation is applicable only rarely; on average about one in every 5.5 × 108 molecules gives up a proton to another water molecule, in accordance with the value of the dissociation constant for water under such conditions. It is a crucial part of the uniqueness of water.
Because water may form hydrogen bonds with solute proton donors and acceptors, it may competitively inhibit the formation of solute intermolecular or intramolecular hydrogen bonds. Consequently, hydrogen bonds between or within solute molecules dissolved in water are almost always unfavorable relative to hydrogen bonds between water and the donors and acceptors for hydrogen bonds on those solutes.[41] Hydrogen bonds between water molecules have an average lifetime of 10−11 seconds, or 10 picoseconds.[42]
Bifurcated and over-coordinated hydrogen bonds in water
A single hydrogen atom can participate in two hydrogen bonds, rather than one. This type of bonding is called "bifurcated" (split in two or "two-forked"). It can exist, for instance, in complex natural or synthetic organic molecules.[43] It has been suggested that a bifurcated hydrogen atom is an essential step in water reorientation.[44]
Acceptor-type hydrogen bonds (terminating on an oxygen's lone pairs) are more likely to form bifurcation (it is called overcoordinated oxygen, OCO) than are donor-type hydrogen bonds, beginning on the same oxygen's hydrogens.[45]
Other liquids
For example, hydrogen fluoride—which has three lone pairs on the F atom but only one H atom—can form only two bonds; (ammonia has the opposite problem: three hydrogen atoms but only one lone pair).
- H−F···H−F···H−F
Further manifestations of solvent hydrogen bonding
- Increase in the melting point, boiling point, solubility, and viscosity of many compounds can be explained by the concept of hydrogen bonding.
- Negative azeotropy of mixtures of HF and water
- The fact that ice is less dense than liquid water is due to a crystal structure stabilized by hydrogen bonds.
- Dramatically higher boiling points of NH3, H2O, and HF compared to the heavier analogues PH3, H2S, and HCl, where hydrogen-bonding is absent.
- Viscosity of anhydrous phosphoric acid and of glycerol
- Dimer formation in carboxylic acids and hexamer formation in hydrogen fluoride, which occur even in the gas phase, resulting in gross deviations from the ideal gas law.
- Pentamer formation of water and alcohols in apolar solvents.
Hydrogen bonds in polymers
Hydrogen bonding plays an important role in determining the three-dimensional structures and the properties adopted by many synthetic and natural proteins. Compared to the C-C, C-O, and C-N bonds that comprise most polymers, hydrogen bonds are far weaker, perhaps 5%. Thus, hydrogen bonds can be broken by chemical or mechanical means while retaining the basic structure of the polymer backbone. This hierarchy of bond strengths (covalent bonds being stronger than hydrogen-bonds being stronger than van der Waals forces) is key to understanding the properties of many materials.[46]
DNA
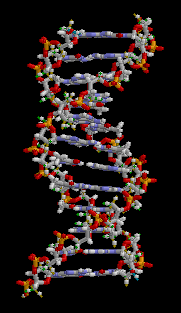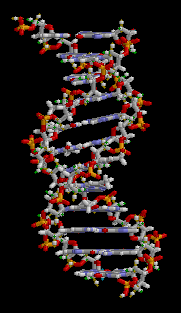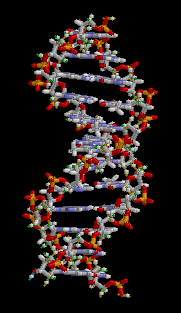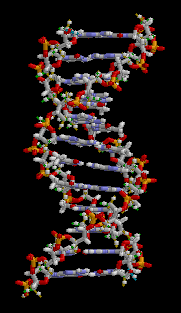
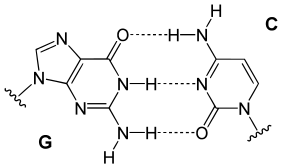
In these macromolecules, bonding between parts of the same macromolecule cause it to fold into a specific shape, which helps determine the molecule's physiological or biochemical role. For example, the double helical structure of DNA is due largely to hydrogen bonding between its base pairs (as well as pi stacking interactions), which link one complementary strand to the other and enable replication.
Proteins
In the secondary structure of proteins, hydrogen bonds form between the backbone oxygens and amide hydrogens. When the spacing of the amino acid residues participating in a hydrogen bond occurs regularly between positions i and i + 4, an alpha helix is formed. When the spacing is less, between positions i and i + 3, then a 310 helix is formed. When two strands are joined by hydrogen bonds involving alternating residues on each participating strand, a beta sheet is formed. Hydrogen bonds also play a part in forming the tertiary structure of protein through interaction of R-groups. (See also protein folding).
Bifurcated H-bond systems are common in alpha-helical transmembrane proteins between the backbone amide C=O of residue i as the H-bond acceptor and two H-bond donors from residue i+4: the backbone amide N-H and a side-chain hydroxyl or thiol H+. The energy preference of the bifurcated H-bond hydroxyl or thiol system is -3.4 kcal/mol or -2.6 kcal/mol, respectively. This type of bifurcated H-bond provides an intrahelical H-bonding partner for polar side-chains, such as serine, threonine, and cysteine within the hydrophobic membrane environments.[47]
The role of hydrogen bonds in protein folding has also been linked to osmolyte-induced protein stabilization. Protective osmolytes, such as trehalose and sorbitol, shift the protein folding equilibrium toward the folded state, in a concentration dependent manner. While the prevalent explanation for osmolyte action relies on excluded volume effects that are entropic in nature, recent circular dichroism (CD) experiments have shown osmolyte to act through an enthalpic effect.[48] The molecular mechanism for their role in protein stabilization is still not well established, though several mechanisms have been proposed. Recently, computer molecular dynamics simulations suggested that osmolytes stabilize proteins by modifying the hydrogen bonds in the protein hydration layer.[49]
Several studies have shown that hydrogen bonds play an important role for the stability between subunits in multimeric proteins. For example, a study of sorbitol dehydrogenase displayed an important hydrogen bonding network which stabilizes the tetrameric quaternary structure within the mammalian sorbitol dehydrogenase protein family.[50]
A protein backbone hydrogen bond incompletely shielded from water attack is a dehydron. Dehydrons promote the removal of water through proteins or ligand binding. The exogenous dehydration enhances the electrostatic interaction between the amide and carbonyl groups by de-shielding their partial charges. Furthermore, the dehydration stabilizes the hydrogen bond by destabilizing the nonbonded state consisting of dehydrated isolated charges.[51]
Wool, being a protein fibre, is held together by hydrogen bonds, causing wool to recoil when stretched. However, washing at high temperatures can permanently break the hydrogen bonds and a garment may permanently lose its shape.
Cellulose
Hydrogen bonds are important in the structure of cellulose and derived polymers in its many different forms in nature, such as cotton and flax.

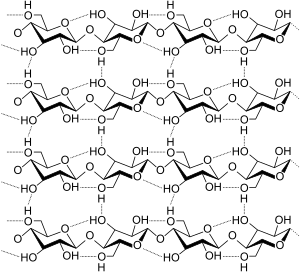
Synthetic polymers
Many polymers are strengthened by hydrogen bonds within and between the chains. Among the synthetic polymers, a well characterized example is nylon, where hydrogen bonds occur in the repeat unit and play a major role in crystallization of the material. The bonds occur between carbonyl and amine groups in the amide repeat unit. They effectively link adjacent chains, which help reinforce the material. The effect is great in aramid fibre, where hydrogen bonds stabilize the linear chains laterally. The chain axes are aligned along the fibre axis, making the fibres extremely stiff and strong.
The hydrogen-bond networks make both natural and synthetic polymers sensitive to humidity levels in the atmosphere because water molecules can diffuse into the surface and disrupt the network. Some polymers are more sensitive than others. Thus nylons are more sensitive than aramids, and nylon 6 more sensitive than nylon-11.
Symmetric hydrogen bond
A symmetric hydrogen bond is a special type of hydrogen bond in which the proton is spaced exactly halfway between two identical atoms. The strength of the bond to each of those atoms is equal. It is an example of a three-center four-electron bond. This type of bond is much stronger than a "normal" hydrogen bond. The effective bond order is 0.5, so its strength is comparable to a covalent bond. It is seen in ice at high pressure, and also in the solid phase of many anhydrous acids such as hydrofluoric acid and formic acid at high pressure. It is also seen in the bifluoride ion [F--H--F]−. Due to severe steric constraint, the protonated form of Proton Sponge (1,8-bis(dimethylamino)naphthalene) and its derivatives also have symmetric hydrogen bonds ([N--H--N]+)[52], although in the case of protonated Proton Sponge, the assembly is bent.[53]
Symmetric hydrogen bonds have been observed recently spectroscopically in formic acid at high pressure (>GPa). Each hydrogen atom forms a partial covalent bond with two atoms rather than one. Symmetric hydrogen bonds have been postulated in ice at high pressure (Ice X). Low-barrier hydrogen bonds form when the distance between two heteroatoms is very small.
Dihydrogen bond
The hydrogen bond can be compared with the closely related dihydrogen bond, which is also an intermolecular bonding interaction involving hydrogen atoms. These structures have been known for some time, and well characterized by crystallography;[54] however, an understanding of their relationship to the conventional hydrogen bond, ionic bond, and covalent bond remains unclear. Generally, the hydrogen bond is characterized by a proton acceptor that is a lone pair of electrons in nonmetallic atoms (most notably in the nitrogen, and chalcogen groups). In some cases, these proton acceptors may be pi-bonds or metal complexes. In the dihydrogen bond, however, a metal hydride serves as a proton acceptor, thus forming a hydrogen-hydrogen interaction. Neutron diffraction has shown that the molecular geometry of these complexes is similar to hydrogen bonds, in that the bond length is very adaptable to the metal complex/hydrogen donor system.[54]
Dynamics probed by spectroscopic means
The dynamics of hydrogen bond structures in water can be probed by the IR spectrum of OH stretching vibration.[55] In the hydrogen bonding network in protic organic ionic plastic crystals (POIPCs), which are a type of phase change material exhibiting solid-solid phase transitions prior to melting, variable-temperature infrared spectroscopy can reveal the temperature dependence of hydrogen bonds and the dynamics of both the anions and the cations.[56] The sudden weakening of hydrogen bonds during the solid-solid phase transition seems to be coupled with the onset of orientational or rotational disorder of the ions.[56]
Application to drugs
Hydrogen bonding is a key to the design of drugs. According to Lipinski's rule of five the majority of orally active drugs tend to have between five and ten hydrogen bonds. These interactions exist between nitrogen–hydrogen and oxygen–hydrogen centers.[57] As with many other rules of thumb, many exceptions exist.
Hydrogen bonding phenomena
- Occurrence of proton tunneling during DNA replication is believed to be responsible for cell mutations.[58]
- High water solubility of many compounds such as ammonia is explained by hydrogen bonding with water molecules.
- Deliquescence of NaOH is caused in part by reaction of OH− with moisture to form hydrogen-bonded H
3O−
2 species. An analogous process happens between NaNH2 and NH3, and between NaF and HF. - The presence of hydrogen bonds can cause an anomaly in the normal succession of states of matter for certain mixtures of chemical compounds as temperature increases or decreases. These compounds can be liquid until a certain temperature, then solid even as the temperature increases, and finally liquid again as the temperature rises over the "anomaly interval"[59]
- Smart rubber utilizes hydrogen bonding as its sole means of bonding, so that it can "heal" when torn, because hydrogen bonding can occur on the fly between two surfaces of the same polymer.
References
- Sweetman, A. M.; Jarvis, S. P.; Sang, Hongqian; Lekkas, I.; Rahe, P.; Wang, Yu; Wang, Jianbo; Champness, N.R.; Kantorovich, L.; Moriarty, P. (2014). "Mapping the force field of a hydrogen-bonded assembly". Nature Communications. 5: 3931. Bibcode:2014NatCo...5.3931S. doi:10.1038/ncomms4931. PMC 4050271. PMID 24875276.
- Hapala, Prokop; Kichin, Georgy; Wagner, Christian; Tautz, F. Stefan; Temirov, Ruslan; Jelínek, Pavel (2014-08-19). "Mechanism of high-resolution STM/AFM imaging with functionalized tips". Physical Review B. 90 (8): 085421. arXiv:1406.3562. Bibcode:2014PhRvB..90h5421H. doi:10.1103/PhysRevB.90.085421.
- Hämäläinen, Sampsa K.; van der Heijden, Nadine; van der Lit, Joost; den Hartog, Stephan; Liljeroth, Peter; Swart, Ingmar (2014-10-31). "Intermolecular Contrast in Atomic Force Microscopy Images without Intermolecular Bonds". Physical Review Letters. 113 (18): 186102. arXiv:1410.1933. Bibcode:2014PhRvL.113r6102H. doi:10.1103/PhysRevLett.113.186102. PMID 25396382. Archived from the original on 2018-01-20. Retrieved 2017-08-30.
- Weinhold, Frank; Klein, Roger A. (2014). "What is a hydrogen bond? Resonance covalency in the supramolecular domain". Chemistry Education Research and Practice. 15: 276–285. doi:10.1039/c4rp00030g.
- Arunan, Elangannan; Desiraju, Gautam R.; Klein, Roger A.; Sadlej, Joanna; Scheiner, Steve; Alkorta, Ibon; Clary, David C.; Crabtree, Robert H.; Dannenberg, Joseph J. (2011-07-08). "Definition of the hydrogen bond (IUPAC Recommendations 2011)". Pure and Applied Chemistry. 83 (8): 1637–1641. doi:10.1351/PAC-REC-10-01-02. ISSN 1365-3075.
- Weinhold, Frank; Klein, Roger A. (2014). "What is a hydrogen bond? Resonance covalency in the supramolecular domain". Chemistry Education Research and Practice. 15: 276–285. doi:10.1039/c4rp00030g.
- Pimentel, G. The Hydrogen Bond Franklin Classics, 2018), ISBN 0343171600
- Jeffrey, G. A.; An introduction to hydrogen bonding; Oxford university press New York, 1997. ISBN 0195095499
- Jeffrey, G. A.; Saenger, W. Hydrogen bonding in biological structures; Springer: Berlin, 1994, 2012 Springer; ISBN 3540579036
- IUPAC, Compendium of Chemical Terminology, 2nd ed. (the "Gold Book") (1997). Online corrected version: (2006–) "hydrogen bond". doi:10.1351/goldbook.H02899
- Steiner, Thomas (2002). "The Hydrogen Bond in the Solid State". Angew. Chem. Int. Ed. 41: 48–76. doi:10.1002/1521-3773(20020104)41:1<48::AID-ANIE48>3.0.CO;2-U.
- Sabin, John R. (1971). "Hydrogen bonds involving sulfur. I. Hydrogen sulfide dimer". J. Am. Chem. Soc. 93 (15): 3613–3620. doi:10.1021/ja00744a012.
- Desiraju, G. R. and Steiner, T. The Weak Hydrogen Bond: In Structural Chemistry and Biology , International Union of Crystallography;2001, ISBN 0198509707
- Nishio, M.; Hirota, M.; Umezawa, Y. The CH–π Interactions; Wiley-VCH, New York, 1998. • Wiley-VCH; 1998) ISBN 0471252905
- Nishio, M (2011). "The CH/[small pi] hydrogen bond in chemistry. "Title". Phys. Chem. Chem. Phys. 13: 13873–13900. doi:10.1039/c1cp20404a.
- Arunan, Elangannan; Desiraju, Gautam R.; Klein, Roger A.; Sadlej, Joanna; Scheiner, Steve; Alkorta, Ibon; Clary, David C.; Crabtree, Robert H.; Dannenberg, Joseph J.; Hobza, Pavel; Kjaergaard, Henrik G.; Legon, Anthony C.; Mennucci, Benedetta; Nesbitt, David J. (2011). "Definition of the hydrogen bond". Pure Appl. Chem. 83 (8): 1637–1641. doi:10.1351/PAC-REC-10-01-02.
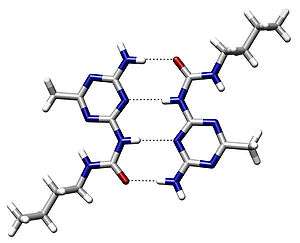
- Beijer, Felix H.; Kooijman, Huub; Spek, Anthony L.; Sijbesma, Rint P.; Meijer, E. W. (1998). "Self-Complementarity Achieved through Quadruple Hydrogen Bonding". Angew. Chem. Int. Ed. 37 (1–2): 75–78. doi:10.1002/(SICI)1521-3773(19980202)37:1/2<75::AID-ANIE75>3.0.CO;2-R.
- Campbell, Neil A.; Brad Williamson; Robin J. Heyden (2006). Biology: Exploring Life. Boston, Massachusetts: Pearson Prentice Hall. ISBN 978-0-13-250882-7. Archived from the original on 2014-11-02.
- Wiley, G.R.; Miller, S.I. (1972). "Thermodynamic parameters for hydrogen bonding of chloroform with Lewis bases in cyclohexane. Proton magnetic resonance study". Journal of the American Chemical Society. 94 (10): 3287. doi:10.1021/ja00765a001.
- Steiner, Thomas (2002). "The Hydrogen Bond in the Solid State". Angew. Chem. Int. Ed. 41: 48–76. doi:10.1002/1521-3773(20020104)41:1<48::AID-ANIE48>3.0.CO;2-U.
- Larson, J. W.; McMahon, T. B. (1984). "Gas-phase bihalide and pseudobihalide ions. An ion cyclotron resonance determination of hydrogen bond energies in XHY- species (X, Y = F, Cl, Br, CN)". Inorganic Chemistry. 23 (14): 2029–2033. doi:10.1021/ic00182a010.
- Emsley, J. (1980). "Very Strong Hydrogen Bonds". Chemical Society Reviews. 9 (1): 91–124. doi:10.1039/cs9800900091.
- V. David, N. Grinberg, S. C. Moldoveanu in Advances in Chromatography Volume 54 (Eds.: E. Grushka, N. Grinberg), CRC Press, Boca Raton, 2018, chapter 3.
- Data obtained using molecular dynamics as detailed in the reference and should be compared to 7.9 kJ/mol for bulk water, obtained using the same calculation.Markovitch, Omer; Agmon, Noam (2007). "Structure and energetics of the hydronium hydration shells" (PDF). J. Phys. Chem. A. 111 (12): 2253–2256. Bibcode:2007JPCA..111.2253M. CiteSeerX 10.1.1.76.9448. doi:10.1021/jp068960g. PMID 17388314. Archived from the original (PDF) on 2014-08-13. Retrieved 2017-10-25.
- Biedermann F, Schneider HJ (May 2016). "Experimental Binding Energies in Supramolecular Complexes". Chemical Reviews. 116 (9): 5216–300. doi:10.1021/acs.chemrev.5b00583. PMID 27136957.
- Legon, A. C.; Millen, D. J. (1987). "Angular geometries and other properties of hydrogen-bonded dimers: a simple electrostatic interpretation of the success of the electron-pair model". Chemical Society Reviews. 16: 467. doi:10.1039/CS9871600467.
- Friebolin, H., "Basic One- and Two- Dimensional NMR Spectroscopy, 4th ed.," VCH: Weinheim, 2008. ISBN 978-3-527-31233-7
- Hobza P, Havlas Z (2000). "Blue-Shifting Hydrogen Bonds". Chem. Rev. 100 (11): 4253–4264. doi:10.1021/cr990050q. PMID 11749346.
- Feldblum, Esther S.; Arkin, Isaiah T. (2014). "Strength of a bifurcated H bond". Proceedings of the National Academy of Sciences. 111 (11): 4085–4090. doi:10.1073/pnas.1319827111. PMC 3964065. PMID 24591597.
- Sun, C. Q.; Sun, Yi (2016). The Attribute of Water: Single Notion, Multiple Myths. ISBN 978-981-10-0178-9.
- Grunenberg, Jörg (2004). "Direct Assessment of Interresidue Forces in Watson−Crick Base Pairs Using Theoretical Compliance Constants". Journal of the American Chemical Society. 126 (50): 16310–1. doi:10.1021/ja046282a. PMID 15600318.
- Isaacs, E.D.; et al. (1999). "Covalency of the Hydrogen Bond in Ice: A Direct X-Ray Measurement". Physical Review Letters. 82 (3): 600–603. Bibcode:1999PhRvL..82..600I. doi:10.1103/PhysRevLett.82.600.
- Ghanty, Tapan K.; Staroverov, Viktor N.; Koren, Patrick R.; Davidson, Ernest R. (2000-02-01). "Is the Hydrogen Bond in Water Dimer and Ice Covalent?". Journal of the American Chemical Society. 122 (6): 1210–1214. doi:10.1021/ja9937019. ISSN 0002-7863.
- Cordier, F; Rogowski, M; Grzesiek, S; Bax, A (1999). "Observation of through-hydrogen-bond (2h)J(HC') in a perdeuterated protein". J Magn Reson. 140 (2): 510–2. Bibcode:1999JMagR.140..510C. doi:10.1006/jmre.1999.1899. PMID 10497060.
- Needham, Paul (2013). "Hydrogen bonding: Homing in on a tricky chemical concept". Studies in History and Philosophy of Science Part A. 44: 51–65. doi:10.1016/j.shpsa.2012.04.001.
- Pauling, L. (1960). The nature of the chemical bond and the structure of molecules and crystals; an introduction to modern structural chemistry (3rd ed.). Ithaca (NY): Cornell University Press. p. 450. ISBN 978-0-8014-0333-0.
- Moore, T. S.; Winmill, T. F. (1912). "The state of amines in aqueous solution". J. Chem. Soc. 101: 1635. doi:10.1039/CT9120101635.
- Latimer, Wendell M.; Rodebush, Worth H. (1920). "Polarity and ionization from the standpoint of the Lewis theory of valence". Journal of the American Chemical Society. 42 (7): 1419–1433. doi:10.1021/ja01452a015.
- Jorgensen, W. L.; Madura, J. D. (1985). "Temperature and size dependence for Monte Carlo simulations of TIP4P water". Mol. Phys. 56 (6): 1381. Bibcode:1985MolPh..56.1381J. doi:10.1080/00268978500103111.
- Zielkiewicz, Jan (2005). "Structural properties of water: Comparison of the SPC, SPCE, TIP4P, and TIP5P models of water". J. Chem. Phys. 123 (10): 104501. Bibcode:2005JChPh.123j4501Z. doi:10.1063/1.2018637. PMID 16178604.
- Jencks, William; Jencks, William P. (1986). "Hydrogen Bonding between Solutes in Aqueous Solution". J. Am. Chem. Soc. 108 (14): 4196. doi:10.1021/ja00274a058.
- Dillon, P. F. (2012). Biophysics: A Physiological Approach. Cambridge University Press. p. 37. ISBN 978-1-139-50462-1.
- Baron, Michel; Giorgi-Renault, Sylviane; Renault, Jean; Mailliet, Patrick; Carré, Daniel; Etienne, Jean (1984). "Hétérocycles à fonction quinone. V. Réaction anormale de la butanedione avec la diamino-1,2 anthraquinone; structure cristalline de la naphto \2,3-f] quinoxalinedione-7,12 obtenue". Can. J. Chem. 62 (3): 526–530. doi:10.1139/v84-087.
- Laage, Damien; Hynes, James T. (2006). "A Molecular Jump Mechanism for Water Reorientation". Science. 311 (5762): 832–5. Bibcode:2006Sci...311..832L. doi:10.1126/science.1122154. PMID 16439623.
- Markovitch, Omer; Agmon, Noam (2008). "The Distribution of Acceptor and Donor Hydrogen-Bonds in Bulk Liquid Water". Molecular Physics. 106 (2): 485. Bibcode:2008MolPh.106..485M. doi:10.1080/00268970701877921.
- Shiao-Wei Kuo (2018). Hydrogen Bonding in Polymer Materials. Wiley-VCH.
- Feldblum, Esther S.; Arkin, Isaiah T. (2014). "Strength of a bifurcated H bond". Proceedings of the National Academy of Sciences. 111 (11): 4085–4090. doi:10.1073/pnas.1319827111. PMC 3964065. PMID 24591597.
- Politi, Regina; Harries, Daniel (2010). "Enthalpically driven peptide stabilization by protective osmolytes". ChemComm. 46 (35): 6449–6451. doi:10.1039/C0CC01763A. PMID 20657920.
- Gilman-Politi, Regina; Harries, Daniel (2011). "Unraveling the Molecular Mechanism of Enthalpy Driven Peptide Folding by Polyol Osmolytes". Journal of Chemical Theory and Computation. 7 (11): 3816–3828. doi:10.1021/ct200455n. PMID 26598272.
- Hellgren, M.; Kaiser, C.; de Haij, S.; Norberg, A.; Höög, J. O. (December 2007). "A hydrogen-bonding network in mammalian sorbitol dehydrogenase stabilizes the tetrameric state and is essential for the catalytic power". Cellular and Molecular Life Sciences. 64 (23): 3129–38. doi:10.1007/s00018-007-7318-1. PMID 17952367.
- Fernández, A.; Rogale K.; Scott Ridgway; Scheraga H. A. (June 2004). "Inhibitor design by wrapping packing defects in HIV-1 proteins". Proceedings of the National Academy of Sciences. 101 (32): 11640–5. Bibcode:2004PNAS..10111640F. doi:10.1073/pnas.0404641101. PMC 511032. PMID 15289598.
- Khashayar Rajabimoghadam Yousef Darwish Umyeena Bashir Dylan Pitman Sidney Eichelberger Maxime A. Siegler Marcel Swart Isaac Garcia-Bosch Aerobic Oxidation of Alcohols by Copper Complexes Bearing Redox-Active Ligands with Tunable H-Bonding https://doi.org/10.1021/jacs.8b08748
- Ozeryanskii, Valery A.; Pozharskii, Alexander F.; Bieńko, Agnieszka J.; Sawka-Dobrowolska, Wanda; Sobczyk, Lucjan (2005-03-01). "[NHN]+ Hydrogen Bonding in Protonated 1,8-Bis(dimethylamino)-2,7-dimethoxynaphthalene. X-ray Diffraction, Infrared, and Theoretical ab Initio and DFT Studies". The Journal of Physical Chemistry A. 109 (8): 1637–1642. doi:10.1021/jp040618l. ISSN 1089-5639. PMID 16833488.
- Crabtree, Robert H.; Siegbahn, Per E. M.; Eisenstein, Odile; Rheingold, Arnold L.; Koetzle, Thomas F. (1996). "A New Intermolecular Interaction: Unconventional Hydrogen Bonds with Element-Hydride Bonds as Proton Acceptor". Acc. Chem. Res. 29 (7): 348–354. doi:10.1021/ar950150s. PMID 19904922.
- Cowan ML; Bruner BD; Huse N; et al. (2005). "Ultrafast memory loss and energy redistribution in the hydrogen bond network of liquid H2O". Nature. 434 (7030): 199–202. Bibcode:2005Natur.434..199C. doi:10.1038/nature03383. PMID 15758995.
- Luo, Jiangshui; Jensen, Annemette H.; Brooks, Neil R.; Sniekers, Jeroen; Knipper, Martin; Aili, David; Li, Qingfeng; Vanroy, Bram; Wübbenhorst, Michael; Yan, Feng; Van Meervelt, Luc; Shao, Zhigang; Fang, Jianhua; Luo, Zheng-Hong; De Vos, Dirk E.; Binnemans, Koen; Fransaer, Jan (2015). "1,2,4-Triazolium perfluorobutanesulfonate as an archetypal pure protic organic ionic plastic crystal electrolyte for all-solid-state fuel cells". Energy & Environmental Science. 8 (4): 1276. doi:10.1039/C4EE02280G.
- Lipinski CA (December 2004). "Lead- and drug-like compounds: the rule-of-five revolution". Drug Discovery Today: Technologies. 1 (4): 337–341. doi:10.1016/j.ddtec.2004.11.007. PMID 24981612.
- Löwdin, P. O. (1963). "Proton Tunneling in DNA and its Biological Implications". Rev. Mod. Phys. 35 (3): 724–732. Bibcode:1963RvMP...35..724L. doi:10.1103/RevModPhys.35.724.
- Law-breaking liquid defies the rules Archived 2011-04-29 at the Wayback Machine. Physicsworld.com (September 24, 2004 )
Further reading
- George A. Jeffrey. An Introduction to Hydrogen Bonding (Topics in Physical Chemistry). Oxford University Press, USA (March 13, 1997). ISBN 0-19-509549-9
External links
- The Bubble Wall (Audio slideshow from the National High Magnetic Field Laboratory explaining cohesion, surface tension and hydrogen bonds)
- isotopic effect on bond dynamics