HtrA serine peptidase 2
Serine protease HTRA2, mitochondrial is an enzyme that in humans is encoded by the HTRA2 gene.[5][6][7] This protein is involved in caspase-dependent apoptosis and in Parkinson's disease.[8][9]
Structure
Gene
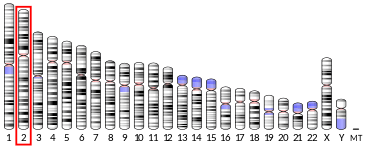
The gene HTRA2 encodes a serine protease. The human gene has 8 exons and locates at chromosome band 2p12.
Protein
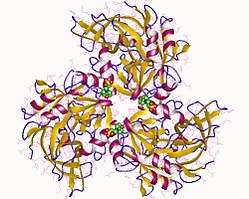
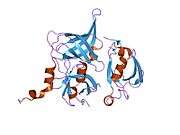
Protein HtrA2, also known as Omi, is a mitochondrially-located serine protease. The human protein Serine protease HTRA2, mitochondrial is 49kDa in size and composed of 458 amino acids. The peptide fragment of 1-31 amino acid is the mitochondrial transition sequence, fragment 32-133 amino acid is propertied, and 134-458 is the mature protein Serine protease HTRA2, mitochondrial, and its theoretical pI of this protein is 6.12.[10] HtrA2 shows similarities with DegS, a bacterial protease present in the periplasm of gram-negative bacteria. Structurally, HtrA2 is a trimeric molecule with central protease domains and a carboxy-terminal PDZ domain, which is characteristic of the HtrA family. The PDZ domain preferentially binds C-terminus of the protein substrate and modulate the proteolytic activity of the trypsin-like protease domain.[11]
Function
The high-temperature requirement (HtrA) family are conserved evolutionarily and these oligomeric serine proteases has been classified in family S1B of the PA protease clan in the MEROPS protease database.[11] The protease activity of the HtrA member HtrA2/Omi is required for mitochondrial homeostasis in mice and humans and inactivating mutations associated with neurodegenerative disorders such as Parkinson's disease.[8] Moreover, HtrA2/Omi is released in the cytosol from the mitochondria during apoptosis and uses its four most N-terminal amino acids to mimic a caspase and be recruited by inhibitor of apoptosis protein (IAP) caspase inhibitors such as XIAP and CIAP1/2. Once bound, the serine protease cleaves the IAP, reducing the cell's inhibition to caspase activation. In summary, HTRA2/Omi contributes to apoptosis through both caspase-dependent and -independent pathways.
Clinical significance
The members of the HtrA family of proteases have been shown playing critical roles in cell physiology and being involved in several pathological processes including cancer[12] and neurodegenerative disease.[11] Strong evidences supported of HtrA2's involvement in oncogenesis. This protein is widely expressed in a variety of cancer cell lines,[13][14][15][16] Analysis of biopsy samples showed changes in expression of HtrA2 in cancer tissues compared with normal tissues.
HtrA2 has recently been identified as a gene related to Parkinson's disease. Mutations in Htra2 have been found in patients suffering from Parkinson's disease. Additionally, mice lacking HtrA2 have a parkinsonian phenotype. This suggests that HtrA2 is linked to Parkinson's disease progression in humans and mice.[9]
Interactions
HtrA serine peptidase 2 has been shown to interact with MAPK14,[5] XIAP[17][18] and BIRC2.[17][18]
References
- GRCh38: Ensembl release 89: ENSG00000115317 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000068329 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Faccio L, Fusco C, Chen A, Martinotti S, Bonventre JV, Zervos AS (Jan 2000). "Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia". The Journal of Biological Chemistry. 275 (4): 2581–8. doi:10.1074/jbc.275.4.2581. PMID 10644717.
- Gray CW, Ward RV, Karran E, Turconi S, Rowles A, Viglienghi D, Southan C, Barton A, Fantom KG, West A, Savopoulos J, Hassan NJ, Clinkenbeard H, Hanning C, Amegadzie B, Davis JB, Dingwall C, Livi GP, Creasy CL (Sep 2000). "Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response". European Journal of Biochemistry / FEBS. 267 (18): 5699–710. doi:10.1046/j.1432-1327.2000.01589.x. PMID 10971580.
- "Entrez Gene: HTRA2 HtrA serine peptidase 2".
- Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, Davies E, Hajnóczky G, Saunders TL, Van Keuren ML, Fernandes-Alnemri T, Meisler MH, Alnemri ES (Oct 2003). "Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice" (PDF). Nature. 425 (6959): 721–7. doi:10.1038/nature02052. hdl:2027.42/62561. PMID 14534547.
- Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D, Gasser T, Wszolek Z, Müller T, Bornemann A, Wolburg H, Downward J, Riess O, Schulz JB, Krüger R (Aug 2005). "Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease". Human Molecular Genetics. 14 (15): 2099–111. doi:10.1093/hmg/ddi215. PMID 15961413.
- "Uniprot: O43464 - HTRA2_HUMAN".
- Vande Walle L, Lamkanfi M, Vandenabeele P (Mar 2008). "The mitochondrial serine protease HtrA2/Omi: an overview". Cell Death and Differentiation. 15 (3): 453–60. doi:10.1038/sj.cdd.4402291. PMID 18174901.
- Bulteau AL, Bayot A (Jun 2011). "Mitochondrial proteases and cancer". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1807 (6): 595–601. doi:10.1016/j.bbabio.2010.12.011. PMID 21194520.
- Bowden MA, Di Nezza-Cossens LA, Jobling T, Salamonsen LA, Nie G (Oct 2006). "Serine proteases HTRA1 and HTRA3 are down-regulated with increasing grades of human endometrial cancer". Gynecologic Oncology. 103 (1): 253–60. doi:10.1016/j.ygyno.2006.03.006. PMID 16650464.
- Lee SH, Lee JW, Kim HS, Kim SY, Park WS, Kim SH, Lee JY, Yoo NJ (May 2003). "Immunohistochemical analysis of Omi/HtrA2 expression in stomach cancer". APMIS. 111 (5): 586–90. doi:10.1034/j.1600-0463.2003.1110508.x. PMID 12887511.
- Narkiewicz J, Klasa-Mazurkiewicz D, Zurawa-Janicka D, Skorko-Glonek J, Emerich J, Lipinska B (May 2008). "Changes in mRNA and protein levels of human HtrA1, HtrA2 and HtrA3 in ovarian cancer". Clinical Biochemistry. 41 (7–8): 561–9. doi:10.1016/j.clinbiochem.2008.01.004. PMID 18241672.
- Zurawa-Janicka D, Kobiela J, Stefaniak T, Wozniak A, Narkiewicz J, Wozniak M, Limon J, Lipinska B (2008). "Changes in expression of serine proteases HtrA1 and HtrA2 during estrogen-induced oxidative stress and nephrocarcinogenesis in male Syrian hamster". Acta Biochimica Polonica. 55 (1): 9–19. doi:10.18388/abp.2008_3123. PMID 18231652.
- Hegde R, Srinivasula SM, Datta P, Madesh M, Wassell R, Zhang Z, Cheong N, Nejmeh J, Fernandes-Alnemri T, Hoshino S, Alnemri ES (Oct 2003). "The polypeptide chain-releasing factor GSPT1/eRF3 is proteolytically processed into an IAP-binding protein". The Journal of Biological Chemistry. 278 (40): 38699–706. doi:10.1074/jbc.M303179200. PMID 12865429.
- Verhagen AM, Silke J, Ekert PG, Pakusch M, Kaufmann H, Connolly LM, Day CL, Tikoo A, Burke R, Wrobel C, Moritz RL, Simpson RJ, Vaux DL (Jan 2002). "HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins". The Journal of Biological Chemistry. 277 (1): 445–54. doi:10.1074/jbc.M109891200. PMID 11604410.
Further reading
- Zurawa-Janicka D, Narkiewicz J, Lipińska B (2007). "[Characterization of the HtrA family of proteins]". Postepy Biochemii. 53 (1): 27–36. PMID 17718385.
- Maruyama K, Sugano S (Jan 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (Oct 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Savopoulos JW, Carter PS, Turconi S, Pettman GR, Karran EH, Gray CW, Ward RV, Jenkins O, Creasy CL (Jul 2000). "Expression, purification, and functional analysis of the human serine protease HtrA2". Protein Expression and Purification. 19 (2): 227–34. doi:10.1006/prep.2000.1240. PMID 10873535.
- Faccio L, Fusco C, Viel A, Zervos AS (Sep 2000). "Tissue-specific splicing of Omi stress-regulated endoprotease leads to an inactive protease with a modified PDZ motif". Genomics. 68 (3): 343–7. doi:10.1006/geno.2000.6263. PMID 10995577.
- Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R (Sep 2001). "A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death". Molecular Cell. 8 (3): 613–21. doi:10.1016/S1097-2765(01)00341-0. PMID 11583623.
- Verhagen AM, Silke J, Ekert PG, Pakusch M, Kaufmann H, Connolly LM, Day CL, Tikoo A, Burke R, Wrobel C, Moritz RL, Simpson RJ, Vaux DL (Jan 2002). "HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins". The Journal of Biological Chemistry. 277 (1): 445–54. doi:10.1074/jbc.M109891200. PMID 11604410.
- Hegde R, Srinivasula SM, Zhang Z, Wassell R, Mukattash R, Cilenti L, DuBois G, Lazebnik Y, Zervos AS, Fernandes-Alnemri T, Alnemri ES (Jan 2002). "Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction". The Journal of Biological Chemistry. 277 (1): 432–8. doi:10.1074/jbc.M109721200. PMID 11606597.
- Vucic D, Deshayes K, Ackerly H, Pisabarro MT, Kadkhodayan S, Fairbrother WJ, Dixit VM (Apr 2002). "SMAC negatively regulates the anti-apoptotic activity of melanoma inhibitor of apoptosis (ML-IAP)". The Journal of Biological Chemistry. 277 (14): 12275–9. doi:10.1074/jbc.M112045200. PMID 11801603.
- van Loo G, van Gurp M, Depuydt B, Srinivasula SM, Rodriguez I, Alnemri ES, Gevaert K, Vandekerckhove J, Declercq W, Vandenabeele P (Jan 2002). "The serine protease Omi/HtrA2 is released from mitochondria during apoptosis. Omi interacts with caspase-inhibitor XIAP and induces enhanced caspase activity". Cell Death and Differentiation. 9 (1): 20–6. doi:10.1038/sj.cdd.4400970. PMID 11803371.
- Li W, Srinivasula SM, Chai J, Li P, Wu JW, Zhang Z, Alnemri ES, Shi Y (Jun 2002). "Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi". Nature Structural Biology. 9 (6): 436–41. doi:10.1038/nsb795. PMID 11967569.
- Yang QH, Church-Hajduk R, Ren J, Newton ML, Du C (Jun 2003). "Omi/HtrA2 catalytic cleavage of inhibitor of apoptosis (IAP) irreversibly inactivates IAPs and facilitates caspase activity in apoptosis". Genes & Development. 17 (12): 1487–96. doi:10.1101/gad.1097903. PMC 196079. PMID 12815069.
- Srinivasula SM, Gupta S, Datta P, Zhang Z, Hegde R, Cheong N, Fernandes-Alnemri T, Alnemri ES (Aug 2003). "Inhibitor of apoptosis proteins are substrates for the mitochondrial serine protease Omi/HtrA2". The Journal of Biological Chemistry. 278 (34): 31469–72. doi:10.1074/jbc.C300240200. PMID 12835328.
- Suzuki Y, Takahashi-Niki K, Akagi T, Hashikawa T, Takahashi R (Feb 2004). "Mitochondrial protease Omi/HtrA2 enhances caspase activation through multiple pathways". Cell Death and Differentiation. 11 (2): 208–16. doi:10.1038/sj.cdd.4401343. PMID 14605674.
- Okada M, Adachi S, Imai T, Watanabe K, Toyokuni SY, Ueno M, Zervos AS, Kroemer G, Nakahata T (Mar 2004). "A novel mechanism for imatinib mesylate-induced cell death of BCR-ABL-positive human leukemic cells: caspase-independent, necrosis-like programmed cell death mediated by serine protease activity". Blood. 103 (6): 2299–307. doi:10.1182/blood-2003-05-1605. PMID 14645012.
- Park HJ, Seong YM, Choi JY, Kang S, Rhim H (Feb 2004). "Alzheimer's disease-associated amyloid beta interacts with the human serine protease HtrA2/Omi". Neuroscience Letters. 357 (1): 63–7. doi:10.1016/j.neulet.2003.11.068. PMID 15036614.
- Guo Y, Cheong N, Zhang Z, De Rose R, Deng Y, Farber SA, Fernandes-Alnemri T, Alnemri ES (Jun 2004). "Tim50, a component of the mitochondrial translocator, regulates mitochondrial integrity and cell death". The Journal of Biological Chemistry. 279 (23): 24813–25. doi:10.1074/jbc.M402049200. PMID 15044455.