FMR1
FMR1 (fragile X mental retardation 1) is a human gene[5] that codes for a protein called fragile X mental retardation protein, or FMRP.[6] This protein, most commonly found in the brain, is essential for normal cognitive development and female reproductive function. Mutations of this gene can lead to fragile X syndrome, intellectual disability, premature ovarian failure, autism, Parkinson's disease, developmental delays and other cognitive deficits.[7] The FMR1 premutation is associated with a wide spectrum of clinical phenotypes that affect more than two million people worldwide.[8]

Function
Synaptic plasticity
FMRP has a diverse array of functions throughout different areas of the neuron; however these functions have not been fully characterized. FMRP has been suggested to play roles in nucleocytoplasmic shuttling of mRNA, dendritic mRNA localization, and synaptic protein synthesis.[9] Studies of Fragile X syndrome have significantly aided in the understanding of the functionality of FMRP through the observed effects of FMRP loss on neurons. A mouse model of fragile X mental retardation implicated the involvement of FMRP in synaptic plasticity.[10] Synaptic plasticity requires the production of new proteins in response to activation of synaptic receptors. It is the production of proteins in response to stimulation which is hypothesized to allow for the permanent physical changes and altered synaptic connections that are linked with the processes of learning and memory.
Group 1 metabotropic glutamate receptor (mGluR) signaling has been implicated in playing an important role in FMRP-dependent synaptic plasticity. Post-synaptic mGluR stimulation results in the up-regulation of protein synthesis through a second messenger system.[11] A role for mGluR in synaptic plasticity is further evidenced by the observation of dendritic spine elongation following mGluR stimulation.[12] Furthermore, mGluR activation results in the synthesis of FMRP near synapses. The produced FMRP associates with polyribosomal complexes after mGluR stimulation, proposing the involvement of fragile X mental retardation protein in the process of translation. This further advocates a role for FMRP in synaptic protein synthesis and the growth of synaptic connections.[13] The loss of FMRP results in an abnormal dendritic spine phenotype. Specifically, deletion of the FMR1 gene in a sample of mice resulted in an increase in spine synapse number.[14]
Role in translation
The proposed mechanism of FMRP's effect upon synaptic plasticity are through its role as a negative regulator of translation. FMRP is an RNA-binding protein which associates with polyribosomes.[13][15] The RNA-binding abilities of FMRP are dependent upon its KH domains and RGG boxes. The KH domain is a conserved motif which characterizes many RNA-binding proteins. Mutagenesis of this domain resulted in impaired FMRP binding to RNA.[16]
FMRP has been shown to inhibit translation of mRNA. Mutation of the FMRP protein resulted in the inability to repress translation as opposed to the wild-type counterpart which was able to do so.[17] As previously mentioned, mGluR stimulation is associated with increased FMRP protein levels. In addition, mGluR stimulation results in increased levels of FMRP target mRNAs. A study found basal levels of proteins encoded by these target mRNAs to be significantly elevated and improperly regulated in FMRP deficient mice.[18]
FMRP translation repression acts by inhibiting the initiation of translation. FMRP directly binds CYFIP1, which in turn binds the translation initiation factor eIF4E. The FMRP-CYFIP1 complex prohibits eIF4E-dependent initiation, thereby acting to repress translation.[19] When applied to the observed phenotype in fragile X Syndrome, the excess protein levels and reduction of translational control can be explained by the loss of translational repression by FMRP in fragile X syndrome.[19][20] FMRP acts to control translation of a large group of target mRNAs; however the extent of FMRPs translational control is unknown. The protein has been shown to repress the translation of target mRNAs at synapses, including those encoding the cytoskeletal proteins Arc/Arg3.1 and MAP1B, and the CaM kinase II.[21] In addition, FMRP binds PSD-95 and GluR1/2 mRNAs. Importantly, these FMRP-binding mRNAs play significant roles in neuronal plasticity.
FMRP translational control has been shown to be regulated by mGluR signaling. mGluR stimulation may result in the transportation of mRNA complexes to synapses for local protein synthesis. FMRP granules have been shown to localize with MAP1B mRNA and ribosomal RNA in dendrites, suggesting this complex as a whole may need to be transported to dendrites for local protein synthesis. In addition, microtubules were found to be a necessary component for the mGluR-dependent translocation of FMRP into dendrites.[9] FMRP may play an additional role in local protein synthesis by aiding in the association of mRNA cargo and microtubules.[22] Thus, FMRP is able to regulate transport efficacy, as well as repression of translation during transport. Finally, FMRP synthesis, ubiquitination, and proteolysis occur rapidly in response to mGluR signaling, suggesting an extremely dynamic role of the translational regulator.[18]
Gene expression
The FMR1 gene is located on the X chromosome and contains a repeated CGG trinucleotide. In most people, the CGG segment is repeated approximately 5-44 times. Higher numbers of repeats of the CGG segment are associated with impaired cognitive and reproductive function. If a person has 45-54 repeats this is considered the “gray zone” or borderline risk, 55-200 repeats is called premutation, and more than 200 repeats is considered a full mutation of the FMR1 gene according to the American College of Medical Genetics and Genomics.[23] The first complete DNA sequence of the repeat expansion in someone with the full mutation was generated by scientists in 2012 using SMRT sequencing.[24] This is an example of a Trinucleotide repeat disorder. Trinucleotide repeat expansion is likely a consequence of strand slippage either during DNA repair or DNA replication.[25]
FMR1 is a chromatin-binding protein that functions in the DNA damage response.[26][27] FMR1 occupies sites on meiotic chromosomes and regulates the dynamics of the DNA damage response machinery during spermatogenesis.[26]
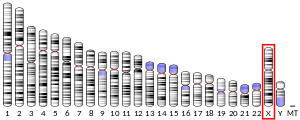
The FMR1 gene can be found on the long (q) arm of the X chromosome at position 27.3, from base pair 146,699,054 to base pair 146,738,156
Related conditions
Fragile X syndrome
Almost all cases of fragile X syndrome are caused by expansion of the CGG trinucleotide repeat in the FMR1 gene. In these cases, CGG is abnormally repeated from 200 to more than 1,000 times. As a result, this part of the FMR1 gene is methylated, which silences the gene (it is turned off and does not make any protein). Without adequate FMR1, severe learning disabilities or intellectual disabilities can develop, along with physical abnormalities seen in fragile X syndrome.
Fewer than 1% of all cases of fragile X syndrome are caused by mutations that delete part or all of the FMR1 gene, or change a base pair, leading to a change in one of the amino acids in the gene. These mutations disrupt the 3-dimensional shape of FMRP or prevent the protein from being synthesized, leading to the signs and symptoms of fragile X syndrome.
A CGG sequence in the FMR1 gene that is repeated between 55 and 200 times is described as a premutation. Although most individuals with the premutation are intellectually normal, some of these individuals have mild versions of the physical features seen in fragile X syndrome (such as prominent ears) and may experience mental health problems such as anxiety or depression.
Fragile X-associated tremor/ataxia syndrome
Premutations are associated with an increased risk of fragile X-associated tremor/ataxia syndrome (FXTAS). FXTAS is characterized by ataxia (loss of coordination), tremor, memory loss, loss of sensation in the lower extremities (peripheral neuropathy) and mental and behavioral changes. The disorder usually develops late in life.
Premature ovarian aging
The FMR1 gene plays a very important role in ovarian function, independent from cognitive/neurological effects. Minor expansions of CGG repeats that do not cause fragile X syndrome are associated with an increased risk for premature ovarian aging, also called occult primary ovarian insufficiency, a condition in which women prematurely deplete their ovarian function.[28][29][30]
Polycystic ovarian syndrome
A very specific sub-genotype of FMR1 has been found to be associated with polycystic ovarian syndrome (PCOS). The gene expression, called heterozygous-normal/low may cause PCOS-like excessive follicle activity and hyperactive ovarian function when women are younger.
Interactions
FMR1 has been shown to interact with:
References
- GRCh38: Ensembl release 89: ENSG00000102081 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000000838 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP (May 1991). "Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome". Cell. 65 (5): 905–14. doi:10.1016/0092-8674(91)90397-H. PMID 1710175.
- Verheij C, Bakker CE, de Graaff E, Keulemans J, Willemsen R, Verkerk AJ, Galjaard H, Reuser AJ, Hoogeveen AT, Oostra BA (June 1993). "Characterization and localization of the FMR-1 gene product associated with fragile X syndrome". Nature. 363 (6431): 722–4. doi:10.1038/363722a0. PMID 8515814.
- "Fragile X Mental Retardation" The Human Gene Compendium
- Milà M, Rodriguez-Revenga L, Matilla-Dueñas A (October 2016). "FMR1 Premutation: Basic Mechanisms and Clinical Involvement". Cerebellum. 15 (5): 543–5. doi:10.1007/s12311-016-0808-7. PMID 27338822.
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ (August 2005). "Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons". Genes, Brain, and Behavior. 4 (6): 350–9. doi:10.1111/j.1601-183X.2005.00128.x. PMID 16098134.
- Huber KM, Gallagher SM, Warren ST, Bear MF (May 2002). "Altered synaptic plasticity in a mouse model of fragile X mental retardation". Proceedings of the National Academy of Sciences of the United States of America. 99 (11): 7746–50. doi:10.1073/pnas.122205699. PMC 124340. PMID 12032354.
- Weiler IJ, Greenough WT (August 1993). "Metabotropic glutamate receptors trigger postsynaptic protein synthesis". Proceedings of the National Academy of Sciences of the United States of America. 90 (15): 7168–71. doi:10.1073/pnas.90.15.7168. PMC 47097. PMID 8102206.
- Vanderklish PW, Edelman GM (February 2002). "Dendritic spines elongate after stimulation of group 1 metabotropic glutamate receptors in cultured hippocampal neurons". Proceedings of the National Academy of Sciences of the United States of America. 99 (3): 1639–44. doi:10.1073/pnas.032681099. PMC 122243. PMID 11818568.
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT (May 1997). "Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation". Proceedings of the National Academy of Sciences of the United States of America. 94 (10): 5395–400. doi:10.1073/pnas.94.10.5395. PMC 24689. PMID 9144248.
- Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ (2006). "Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses". Molecular and Cellular Neurosciences. 32 (1–2): 37–48. doi:10.1016/j.mcn.2006.02.001. PMID 16631377.
- Brown V, Small K, Lakkis L, Feng Y, Gunter C, Wilkinson KD, Warren ST (June 1998). "Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein". The Journal of Biological Chemistry. 273 (25): 15521–7. doi:10.1074/jbc.273.25.15521. PMID 9624140.
- Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G (April 1994). "Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome". Cell. 77 (1): 33–9. doi:10.1016/0092-8674(94)90232-1. PMID 8156595.
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U (February 2001). "Evidence that fragile X mental retardation protein is a negative regulator of translation". Human Molecular Genetics. 10 (4): 329–38. doi:10.1093/hmg/10.4.329. PMID 11157796.
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E (August 2006). "Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression". Neuron. 51 (4): 441–54. doi:10.1016/j.neuron.2006.07.005. PMID 16908410.
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C (September 2008). "The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP". Cell. 134 (6): 1042–54. doi:10.1016/j.cell.2008.07.031. PMID 18805096.
- Muddashetty RS, Kelić S, Gross C, Xu M, Bassell GJ (May 2007). "Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome". The Journal of Neuroscience. 27 (20): 5338–48. doi:10.1523/JNEUROSCI.0937-07.2007. PMID 17507556.
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C (February 2003). "The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses". Cell. 112 (3): 317–27. doi:10.1016/S0092-8674(03)00079-5. PMID 12581522.
- Estes PS, O'Shea M, Clasen S, Zarnescu DC (October 2008). "Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons". Molecular and Cellular Neurosciences. 39 (2): 170–9. doi:10.1016/j.mcn.2008.06.012. PMID 18655836.
- "Technical Standards and Guidelines for Fragile X". American College of Medical Genetics. 2000-10-02. Retrieved 2013-03-29.
- Loomis EW, Eid JS, Peluso P, Yin J, Hickey L, Rank D, McCalmon S, Hagerman RJ, Tassone F, Hagerman PJ (January 2013). "Sequencing the unsequenceable: expanded CGG-repeat alleles of the fragile X gene". Genome Research. 23 (1): 121–8. doi:10.1101/gr.141705.112. PMC 3530672. PMID 23064752.
- Usdin K, House NC, Freudenreich CH (2015). "Repeat instability during DNA repair: Insights from model systems". Crit. Rev. Biochem. Mol. Biol. 50 (2): 142–67. doi:10.3109/10409238.2014.999192. PMC 4454471. PMID 25608779.
- Alpatov R, Lesch BJ, Nakamoto-Kinoshita M, Blanco A, Chen S, Stützer A, Armache KJ, Simon MD, Xu C, Ali M, Murn J, Prisic S, Kutateladze TG, Vakoc CR, Min J, Kingston RE, Fischle W, Warren ST, Page DC, Shi Y (May 2014). "A chromatin-dependent role of the fragile X mental retardation protein FMRP in the DNA damage response". Cell. 157 (4): 869–81. doi:10.1016/j.cell.2014.03.040. PMC 4038154. PMID 24813610.
- Dockendorff TC, Labrador M (May 2018). "The Fragile X Protein and Genome Function". Mol. Neurobiol. 56 (1): 711–721. doi:10.1007/s12035-018-1122-9. PMID 29796988.
- Gleicher N, Barad DH (August 2010). "The FMR1 gene as regulator of ovarian recruitment and ovarian reserve". Obstetrical & Gynecological Survey. 65 (8): 523–30. doi:10.1097/OGX.0b013e3181f8bdda. PMID 20955631.
- Chatterjee S, Maitra A, Kadam S, Patel Z, Gokral J, Meherji P (August 2009). "CGG repeat sizing in the FMR1 gene in Indian women with premature ovarian failure". Reproductive Biomedicine Online. 19 (2): 281–6. doi:10.1016/s1472-6483(10)60086-7. PMID 19712568.
- Streuli I, Fraisse T, Ibecheole V, Moix I, Morris MA, de Ziegler D (August 2009). "Intermediate and premutation FMR1 alleles in women with occult primary ovarian insufficiency". Fertility and Sterility. 92 (2): 464–70. doi:10.1016/j.fertnstert.2008.07.007. PMID 18973899.
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL (July 2001). "A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P". Proceedings of the National Academy of Sciences of the United States of America. 98 (15): 8844–9. doi:10.1073/pnas.151231598. PMC 37523. PMID 11438699.
- Bardoni B, Castets M, Huot ME, Schenck A, Adinolfi S, Corbin F, Pastore A, Khandjian EW, Mandel JL (July 2003). "82-FIP, a novel FMRP (fragile X mental retardation protein) interacting protein, shows a cell cycle-dependent intracellular localization". Human Molecular Genetics. 12 (14): 1689–98. doi:10.1093/hmg/ddg181. PMID 12837692.
- Siomi MC, Zhang Y, Siomi H, Dreyfuss G (July 1996). "Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them". Molecular and Cellular Biology. 16 (7): 3825–32. doi:10.1128/mcb.16.7.3825. PMC 231379. PMID 8668200.
- Zhang Y, O'Connor JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL, Dreyfuss G (November 1995). "The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2". The EMBO Journal. 14 (21): 5358–66. doi:10.1002/j.1460-2075.1995.tb00220.x. PMC 394645. PMID 7489725.
- Ceman S, Brown V, Warren ST (December 1999). "Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex". Molecular and Cellular Biology. 19 (12): 7925–32. doi:10.1128/mcb.19.12.7925. PMC 84877. PMID 10567518.
- Bardoni B, Schenck A, Mandel JL (December 1999). "A novel RNA-binding nuclear protein that interacts with the fragile X mental retardation (FMR1) protein". Human Molecular Genetics. 8 (13): 2557–66. doi:10.1093/hmg/8.13.2557. PMID 10556305.
Further reading
- Bassell GJ, Warren ST (October 2008). "Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function". Neuron. 60 (2): 201–14. doi:10.1016/j.neuron.2008.10.004. PMC 3691995. PMID 18957214.
- Hagerman PJ, Hagerman RJ (May 2004). "The fragile-X premutation: a maturing perspective". American Journal of Human Genetics. 74 (5): 805–16. doi:10.1086/386296. PMC 1181976. PMID 15052536.
- Hagerman RJ, Leavitt BR, Farzin F, Jacquemont S, Greco CM, Brunberg JA, Tassone F, Hessl D, Harris SW, Zhang L, Jardini T, Gane LW, Ferranti J, Ruiz L, Leehey MA, Grigsby J, Hagerman PJ (May 2004). "Fragile-X-associated tremor/ataxia syndrome (FXTAS) in females with the FMR1 premutation". American Journal of Human Genetics. 74 (5): 1051–6. doi:10.1086/420700. PMC 1181968. PMID 15065016.
- Jacquemont S, Hagerman RJ, Leehey MA, Hall DA, Levine RA, Brunberg JA, Zhang L, Jardini T, Gane LW, Harris SW, Herman K, Grigsby J, Greco CM, Berry-Kravis E, Tassone F, Hagerman PJ (January 2004). "Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population". JAMA. 291 (4): 460–9. doi:10.1001/jama.291.4.460. PMID 14747503.
- Jin P, Alisch RS, Warren ST (November 2004). "RNA and microRNAs in fragile X mental retardation". Nature Cell Biology. 6 (11): 1048–53. doi:10.1038/ncb1104-1048. PMID 15516998.
- Jin P, Warren ST (March 2003). "New insights into fragile X syndrome: from molecules to neurobehaviors". Trends in Biochemical Sciences. 28 (3): 152–8. CiteSeerX 10.1.1.532.4162. doi:10.1016/S0968-0004(03)00033-1. PMID 12633995.
- O'Donnell WT, Warren ST (2002). "A decade of molecular studies of fragile X syndrome". Annual Review of Neuroscience. 25: 315–38. doi:10.1146/annurev.neuro.25.112701.142909. PMID 12052912.
- Oostra BA, Chiurazzi P (December 2001). "The fragile X gene and its function". Clinical Genetics. 60 (6): 399–408. doi:10.1034/j.1399-0004.2001.600601.x. PMID 11846731.
- Oostra BA, Willemsen R (October 2003). "A fragile balance: FMR1 expression levels". Human Molecular Genetics. 12 Spec No 2 (90002): R249–57. doi:10.1093/hmg/ddg298. PMID 12952862.
- Nicola NA, Metcalf D (October 1991). "Subunit promiscuity among hemopoietic growth factor receptors". Cell. 67 (1): 1–4. doi:10.1016/0092-8674(91)90564-F. PMID 1913811.
- Sielska D, Milewski M, Bal J (2003). "[Molecular pathogenesis of fragile X syndrome]". Medycyna Wieku Rozwojowego. 6 (4): 295–308. PMID 12810982.
- Bagni C, Greenough WT (May 2005). "From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome". Nature Reviews. Neuroscience. 6 (5): 376–87. doi:10.1038/nrn1667. PMID 15861180.
- Huber KM (April 2006). "The fragile X-cerebellum connection". Trends in Neurosciences. 29 (4): 183–5. doi:10.1016/j.tins.2006.02.001. PMID 16500716.
- Loesch DZ, Bui QM, Dissanayake C, Clifford S, Gould E, Bulhak-Paterson D, Tassone F, Taylor AK, Hessl D, Hagerman R, Huggins RM (2007). "Molecular and cognitive predictors of the continuum of autistic behaviours in fragile X". Neuroscience and Biobehavioral Reviews. 31 (3): 315–26. doi:10.1016/j.neubiorev.2006.09.007. PMC 2145511. PMID 17097142.
External links
| Wikimedia Commons has media related to Fragile X mental retardation protein. |