Conformational proofreading
Conformational proofreading or conformational selection is a general mechanism of molecular recognition systems in which introducing a structural mismatch between a molecular recognizer and its target, or an energetic barrier, enhances the recognition specificity and quality.[1][2][3][4][5][6] Conformational proofreading does not require the consumption of energy and may therefore be used in any molecular recognition system. Conformational proofreading is especially useful in scenarios where the recognizer has to select the appropriate target among many similar competitors.
Balancing correct and incorrect binding
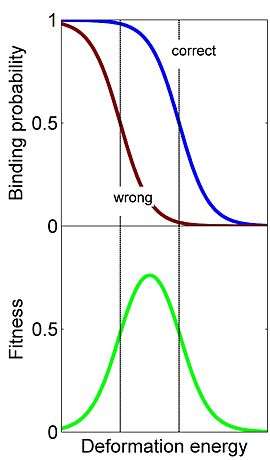
Molecular recognition takes place in a noisy, crowded biological environment and the recognizer often has to cope with the task of selecting its target among a variety of similar competitors. For example, the ribosome has to select the correct tRNA that matches the mRNA codon among many structurally similar tRNAs. If the recognizer and its correct target match perfectly like a lock and a key, then the binding probability will be high since no deformation is required upon binding. At the same time, the recognizer might also bind to a competitor with a similar structure with high probability. Introducing a structural mismatch between the recognizer (lock) and the key reduces the binding probability to the correct target but reduces even more the binding probability to a similar wrong target and thus improves the specificity. Yet, introducing too much deformation drastically reduces binding probability to the correct target. Therefore, the optimal balance between maximizing the correct binding probability and minimizing the incorrect binding probability is achieved when the recognizer is slightly off target. This suggests that conformational changes during molecular recognition processes, such as the induced fit[7] mechanism, are advantageous for enhancing the specificity of recognition.
Use by homologous recombination for homology search
The mechanism of conformational proofreading is utilized in the system of homologous recombination to discern between similar DNA sequences.[3][4] Homologous recombination facilitates the exchange of genetic material between homologous DNA molecules. This crucial process requires detecting a specific homologous DNA sequence within a huge variety of heterologous sequences. The detection is mediated by RecA in E. coli, or members of its superfamily in other organisms. RecA first polymerizes along a stretch of single-stranded DNA, and then this protein-DNA filament searches for homology along double-stranded DNA. In the RecA-DNA filament, the distance between bases increases significantly with respect to the bare 3.4 Å in the double-strand (by 50% on average[8]). This sets a significant energetic barrier on the search, since the double-stranded DNA has to stretch by the same magnitude to check for homology. By formulating the DNA recognition process as a signal detection problem, it was shown that the experimentally observed RecA-induced DNA deformation and the binding energetics are fine-tuned to ensure optimal sequence detection. The amount of deformation is such that binding to homologous DNA sequences only slightly decreases, while binding to wrong sequences decreases significantly. This is exactly the conformational proofreading mechanism.
Experimental evidence for conformational proofreading by homologous recombination
The group of C. Dekker (Delft University) directly probed the interactions involved in homology search by combining magnetic and optical tweezers.[9] They have found that homology search and recognition requires opening of the helix and can therefore be accelerated by unwinding the DNA. This is exactly the energy barrier predicted by the conformational proofreading model. The data indicate a physical picture for homology recognition in which the fidelity of the search process is governed by the distance between the DNA-binding sites. The authors conclude that their interpretation of the measurements "is akin to a conformational proofreading scheme ... where the dsDNA, and not the RecA filament, is the active, recognizing search entity. A large conformational mismatch exists between the target-bound and unbound states of the dsDNA. The target-bound state is accessed via energetically unfavorable intermediate states, as discussed above. The conformational mismatch improves the selectivity of the recognition reaction." In other words, they identified the energetic barrier and have shown that indeed the double-stranded DNA is the active participant, since it has to pass this barrier.
Use by ribosome for tRNA decoding
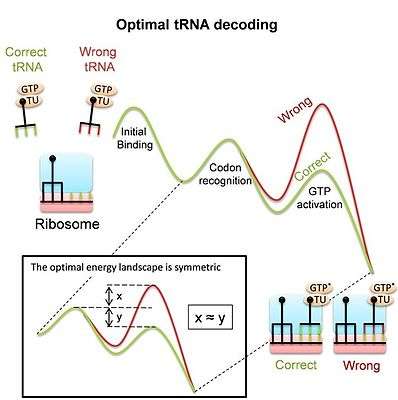
The ribosome is a complex molecular machine that, in order to synthesize proteins during the translation process, has to decode mRNAs by pairing their codons with matching tRNAs. Decoding is a major determinant of fitness and requires accurate and fast selection of correct tRNAs among many similar competitors. One must have in mind that most binding events are by non-matching (“non-cognate”) tRNAs and the ribosome needs to reject those as fast as possible in order to vacate the binding site. At the same time, the ribosome should keep the matching tRNAs bound enough time to allow the protein synthesis process ensue. Despite the importance of tRNA decoding, it was unclear until recently whether the modern ribosome, and in particular its large conformational changes during decoding, are the outcome of adaptation to its task as a decoder or the result of other constraints. Recent study[5] derived the energy landscape that provides optimal discrimination between competing tRNA substrates, and thereby optimal tRNA decoding. The optimal landscape is a symmetric one (see image). The study shows that the measured landscape of the prokaryotic ribosome is indeed symmetric. This model suggests that conformational changes of the ribosome and tRNA during decoding are means to obtain such an optimal tRNA decoder. The fact that both homologous recombination and tRNA decoding utilize conformational proofreading suggests that this is a generic mechanism that may be utilized broadly by molecular recognition systems.
In other biological systems
Human UV-damage repair
A recent study shows that conformational proofreading is used by human DNA repair mechanisms.[10] The research focused on the question of how DNA-repair proteins scan the human genome for UV-induced damage during the initial step of nucleotide excision repair (NER). Detailed single-molecule measurements revealed how the human UV-damaged DNA-binding protein (UV-DDB) performs a 3D search. The authors find that "UV-DDB examines sites on DNA in discrete steps before forming long-lived, nonmotile UV-DDB dimers (DDB1-DDB2)2 at sites of damage. Analysis of the rates of dissociation for the transient binding molecules on both undamaged and damaged DNA show multiple dwell times over three orders of magnitude... These intermediate states are believed to represent discrete UV-DDB conformers on the trajectory to stable damage detection." The authors conclude from their detailed kinetic measurements that UV-DDB recognizes lesions using a conformational proofreading mechanism via multiple intermediates.
Other recognition schemes
Relation to kinetic proofreading
In the kinetic proofreading[11][12] schema, a time delay (equivalently, an irreversible intermediate stage) is introduced during the formation of the correct or incorrect complexes. This time delay reduces the production rates of both complexes but enhances the fidelity beyond the equilibrium limit. The irreversibility of the scheme requires an energy source. The time delay in kinetic proofreading is analogous to the spatial difference in conformational proofreading. However, the conformational proofreading can be an equilibrium scheme that does not consume energy.
References
- Savir Y & Tlusty T (2007). Scalas, Enrico (ed.). "Conformational Proofreading: The Impact of Conformational Changes on the Specificity of Molecular Recognition". PLoS ONE. 2 (5): e468. Bibcode:2007PLoSO...2..468S. doi:10.1371/journal.pone.0000468. PMC 1868595. PMID 17520027.
- Savir Y, Tlusty T (2008). "Optimal Design of a Molecular Recognizer : Molecular Recognition as a Bayesian Signal Detection Problem". IEEE J Sel Topics Signal Process. 2 (3): 390–399. arXiv:1007.4527. Bibcode:2008ISTSP...2..390S. doi:10.1109/JSTSP.2008.923859.
- Savir Y, Tlusty T (2010). "RecA-mediated homology search as a nearly optimal signal detection system". Molecular Cell. 40 (3): 388–96. arXiv:1011.4382. doi:10.1016/j.molcel.2010.10.020. PMID 21070965.
- Rambo RP, Williams GJ, Tainer JA (2010). "Achieving Fidelity in Homologous Recombination Despite Extreme Complexity: Informed Decisions by Molecular Profiling". Molecular Cell. 40 (3): 347–48. doi:10.1016/j.molcel.2010.10.032. PMC 3003302. PMID 21070960.
- Savir, Yonatan; Tlusty, Tsvi (Apr 11, 2013). "The ribosome as an optimal decoder: a lesson in molecular recognition". Cell. 153 (2): 471–9. doi:10.1016/j.cell.2013.03.032. PMID 23582332.
- Alon U (2008). "Journal Club". Nature. 453 (7196): 701. Bibcode:2008Natur.453..701A. doi:10.1038/453701e.
- Koshland, D. E. (1958). "Application of a Theory of Enzyme Specificity to Protein Synthesis". Proc Natl Acad Sci U S A. 44 (2): 98–104. Bibcode:1958PNAS...44...98K. doi:10.1073/pnas.44.2.98. PMC 335371. PMID 16590179.
- Chen Z, Yang H, Pavletich NP (2008). "Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures". Nature. 453 (7194): 489–4. Bibcode:2008Natur.453..489C. doi:10.1038/nature06971. PMID 18497818.
- De Vlaminck I, van Loenhout MT, Zweifel L, den Blanken J, Hooning K, Hage S, Kerssemakers J, Dekker C (2012). "Mechanism of Homology Recognition in DNA Recombination from Dual-Molecule Experiments". Molecular Cell. 46 (5): 616–624. doi:10.1016/j.molcel.2012.03.029. PMID 22560720.
- Ghodke H, Wang H, Hsieh CL, Woldemeskel S, Watkins SC, Rapić-Otrin V, Van Houten B (May 6, 2014). "Single-molecule analysis reveals human UV-damaged DNA-binding protein (UV-DDB) dimerizes on DNA via multiple kinetic intermediates". Proc Natl Acad Sci U S A. 111 (18): 1862–71. Bibcode:2014PNAS..111E1862G. doi:10.1073/pnas.1323856111. PMC 4020048. PMID 24760829.
- Hopfield JJ (1974). "Kinetic Proofreading: A New Mechanism for Reducing Errors in Biosynthetic Processes Requiring High Specificity". Proc Natl Acad Sci U S A. 71 (10): 4135–4139. Bibcode:1974PNAS...71.4135H. doi:10.1073/pnas.71.10.4135. PMC 434344. PMID 4530290.
- Ninio J (1975). "Kinetic amplification of enzyme discrimination Biochimie". Biochimie. 57 (5): 587–595. doi:10.1016/S0300-9084(75)80139-8. PMID 1182215.