Adenylylation
Adenylylation,[1][2] more commonly known as AMPylation, is a process in which an adenosine monophosphate (AMP) molecule is covalently attached to the amino acid side chain of a protein.[3] This covalent addition of AMP to a hydroxyl side chain of the protein is a posttranslational modification.[4] Adenylylation involves a phosphodiester bond between a hydroxyl group of the molecule undergoing adenylylation, and the phosphate group of the adenosine monophosphate nucleotide (i.e. adenylic acid). Enzymes that are capable of catalyzing this process are called AMPylators. The known amino acids to be targeted in the protein are tyrosine and threonine, and sometimes serine[5]. When charges on a protein undergo a change, it affects the characteristics of the protein, normally by altering its shape via interactions of the amino acids which make up the protein. AMPylation can have various effects on the protein. These are properties of the protein like, stability, enzymatic activity, co-factor binding, and many other functional capabilities of a protein. The most commonly identified protein to receive AMPylation are GTPases, and glutamine synthetase.
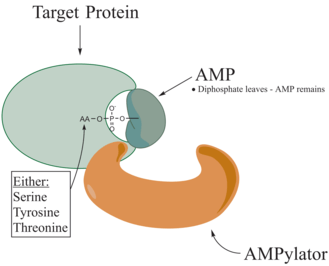
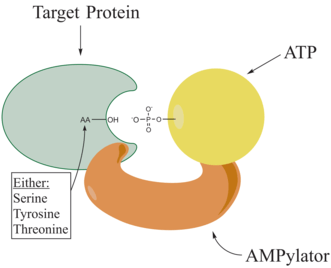
AMPylators
Enzymes responsible for AMPylation, called AMPylators, fall into two different families, all depending on their structural properties and mechanism used. These two families are the DNA-β-polymerase-like and the Fic family.[6]
DNA-β-polymerase-like, is a family of Nucleotidyltransferase[4]. It more specifically is known as the GlnE family. There is a specific motif that is used to clarify this particular family. The motif consists of a three stranded β-sheet which is part of magnesium ion coordination and phosphate binding. Aspartate is essential for the activity to occur in this family.
The Fic family, which is a filamentation induced by cyclic AMP domain, is known to perform AMPylation. This family of proteins are found in all domains of life on earth. It is mediated via a mechanism of ATP-binding-site alpha helix motif. Infectious bacteria use this domain to interrupt phagocytosis and cause cell death. Fic domains are evolutionarily conserved domains in prokaryotes and eukaryotes that belong to the Fido domain superfamily[4].
These enzymes have been shown to be comparable to kinases due to their ATP hydrolysis activity and reversible transfer of the metabolite to a hydroxyl side chain of the protein substrate.
De-AMPylation
GS-ATasE (GlnE) is an AMPylator that has been shown to catalyze de-AMPylation of glutamine synthetase by removing the covalent linkage between AMP and a hydroxyl residue of a protein. It contains two adenylyl transferase domains that are involved in the addition and removal of AMP to glutamine synthetase. De-AMPylation occurs at the N-terminus of the domain. Following the removal of AMP from glutamine synthetase, GS-ATase forms ADP and unmodified glutamine synthetase.[4]
Pathogenicity
Bacteria proteins, also known as effectors, have been shown to use AMPylation. Effectors such as VopS, IbpA, and DrrA, have been shown to AMPylate host GTPases and cause actin cytoskeleton changes. GTPases are common targets of AMPylators. Rho, Rab, and Arf GTPase families are involved in actin cytoskeleton dynamics and vesicular trafficking. They also play roles in cellular control mechanisms such as phagocytosis in the host cell.
The pathogen enhances or prevents its internalization by either inducing or inhibiting host cell phagocytosis[4]. Vibrio parahaemolyticus is a Gram-negative bacterium that causes food poisoning as a result of raw or undercooked seafood consumption in humans.[7] VopS, a type III effector found in Vibrio parahaemolyticus, contains a Fic domain that has a conserved HPFx(D/E)GN(G/K)R motif that contains a histidine residue essential for AMPylation. VopS blocks actin assembly by modifying threonine residue in the switch 1 region of Rho GTPases. The transfer of an AMP moiety using ATP to the threonine residue results in steric hindrance, and thus prevents Rho GTPases from interacting with downstream effectors. As a result, the host cell’s actin cytoskeleton control is disabled, leading to cell rounding.[4][7]
IbpA is secreted into eukaryotic cells from H. somni, a Gram-negative bacterium in cattle that causes respiratory epithelium infection. This effector contains two Fic domains at the C-terminal region. AMPylation of the IbpA Fic domain of Rho family GTPases is responsible for its cytotoxicity. The AMPylation on a tyrosine residue of the switch 1 region blocks the interaction of the GTPases with downstream substrates such as PAK.
References
- Han KK, Martinage A (1992). "Post-translational chemical modification(s) of proteins". Int. J. Biochem. 24 (1): 19–28. PMID 1582530.
- Garrett, R.H., and C.M. Grisham. Biochemistry. 3rd ed. Belmont, CA: Thomas, 2007. 815-20
- Itzen, Aymelt, Wulf Blankenfeldt, and Roger S. Goody. "Adenylylation: renaissance of a forgotten post-translational modification." Trends in Biochemical Sciences 36.4 (2011): 221-228. Print.
- Woolery, Andrew. "AMPylation: something old is new again." Frontiers in Microbiology 1 (2010): 1-18. Print.
- Casey, A. K., & Orth, K. (2018, February 14). Enzymes Involved in AMPylation and deAMPylation. Chemical Reviews. American Chemical Society. https://doi.org/10.1021/acs.chemrev.7b00145
- Hedberg, C., & Itzen, A. (2015). Molecular perspectives on protein adenylylation. ACS Chemical Biology, 10(1), 12–21. https://doi.org/10.1021/cb500854e
- Luong, P., L. N. Kinch, C. A. Brautigam, N. V. Grishin, D. R. Tomchick, and K. Orth. "Kinetic and Structural Insights into the Mechanism of AMPylation by VopS Fic Domain." Journal of Biological Chemistry 285.26 (2010): 20155-20163. Print.