2-dehydro-3-deoxy-phosphogluconate aldolase
In enzymology, a 2-dehydro-3-deoxy-phosphogluconate aldolase (EC 4.1.2.14), commonly known as KDPG aldolase, is an enzyme that catalyzes the chemical reaction
- 2-dehydro-3-deoxy-D-gluconate 6-phosphate pyruvate + D-glyceraldehyde 3-phosphate
| 2-dehydro-3-deoxy-phosphogluconate aldolase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 4.1.2.14 | ||||||||
| CAS number | 9024-53-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Hence, this enzyme primarily has one substrate, 2-dehydro-3-deoxy-D-gluconate 6-phosphate, and two products, pyruvate and D-glyceraldehyde 3-phosphate.
This enzyme belongs to the family of lyases, specifically the aldehyde-lyases, which cleave carbon-carbon bonds. It is used in the Entner–Doudoroff pathway in prokaryotes, feeding into glycolysis. 2-dehydro-3-deoxy-phosphogluconate aldolase is one of the two enzymes distinguishing this pathway from the more commonly known Embden–Meyerhof–Parnas pathway.[1] This enzyme also participates in following 3 metabolic pathways: pentose phosphate pathway, pentose and glucuronate interconversions, and arginine and proline metabolism.
In addition to the cleavage of 2-dehydro-3-deoxy-D-gluconate 6-phosphate, it is also found to naturally catalyze Schiff base formation between a lysine E-amino acid group and carbonyl compounds, decarboxylation of oxaloacetate, and exchange of solvent protons with the methyl hydrogen atoms of pyruvate.[2]
Nomenclature
The systematic name of this enzyme class is 2-dehydro-3-deoxy-D-gluconate-6-phosphate D-glyceraldehyde-3-phosphate-lyase (pyruvate-forming). Other names in common use include:
- KDPG aldolase
- phospho-2-keto-3-deoxygluconate aldolase
- phospho-2-keto-3-deoxygluconic aldolase
- 2-keto-3-deoxy-6-phosphogluconic aldolase
- 2-keto-3-deoxy-6-phosphogluconate aldolase
- 6-phospho-2-keto-3-deoxygluconate aldolase
- ODPG aldolase, 2-oxo-3-deoxy-6-phosphogluconate aldolase
- 2-keto-3-deoxygluconate-6-P-aldolase
- 2-keto-3-deoxygluconate-6-phosphate aldolase
- 2-dehydro-3-deoxy-D-gluconate-6-phosphate
- D-glyceraldehyde-3-phosphate-lyase
Enzyme structure
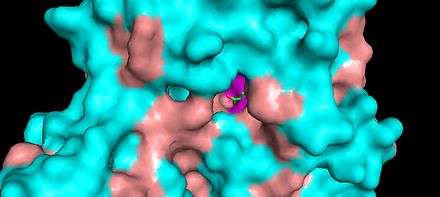
KDPG Aldolase was recently determined to be a trimer through crystallographic three-fold symmetry, with 225 residues.[2][3] The enzyme was determined to have a molecular weight of 23,942.[4] The trimer is stabilized primarily through hydrophobic interactions. The molecule has tertiary folding similar to triosephosphate isomerase and the A-domain of pyruvate kinase, forming an eight strand α/β-barrel structure.[3][5] The α/β-barrel structure is capped on one side by the N-terminal helix. The other side, the carboxylic side, contains the active site.[6] Each subunit contains a phosphate-ion bound in position of the aldolase biding site.[7] It has been found that there are four cysteinyl groups present in each subunit, with two readily accessible and two buried in the subunit.[8]
The active site contains the zwitterionic pair Glu-45/Lys-133.[9] The Lysine, which is involved in the formation of the Schiff base is coordinated by a phosphate ion and two solvent water molecules.[6][7] The first water molecule serves as a shuttle between the Glutamate and the substrate, staying bound to the enzyme throughout the catalytic cycle.[7] The second water molecule is a product of the dehydration of the carbinolamine that leads to the formation of the Schiff base.[7] It also functions as the nucleophile during hydrolysis of the enzyme-product Schiff base, leading to the release of pyruvate.[7]
As of late 2007, 13 structures have been solved for this class of enzymes, with PDB accession codes 1EUA, 1EUN, 1FQ0, 1FWR, 1KGA, 1MXS, 1WA3, 1WAU, 1WBH, 2C0A, 2NUW, 2NUX, and 2NUY.
Enzyme mechanism

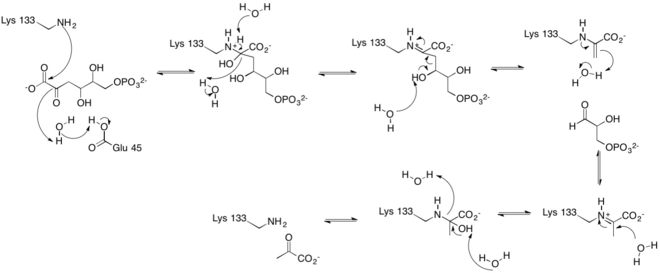
One of the reactions KDPG Aldolase catalyzes, as in the Entner–Doudoroff pathway, is the reversible cleavage of 2-keto-3-deoxy-6-phosphogluconate (KDPG) into pyruvate and D-glyceraldehyde-3-phosphate.[9][10] This occurs through a stereospecific retro-aldol cleavage.[7] A proton transfer between the zwitterionic pair Glu-45/Lys-133 in the active site activates Lysine to serve as the nucleophile in the first step and Glutamate to aid in the base catalysis involved in the carbon-carbon cleavage.[9] Lysine Residue 133 serves as the nucleophile and attacks the carbonyl group of 2-Keto-3-deoxy-6-phosphogluconate to form a protonated carbinolamine intermediate, also known as a Schiff base intermediate.[7][9][10] The intermediate is stabilized by hydrogen bonding with residues in the active site.[9] A three carbon residue, glyceraldehyde 3-phosphate, is cleaved off through base catalysis with a water molecule and residue Glu-45.[7][9] The pyruvate is generated through the nucleophilic attack of water on the Schiff-base to reform a ketone. Aromatic interaction with Phe-135 ensures the stereospecific addition involved in the reverse process.[9]
KDPG aldolase has also been shown to catalyze the exchange of hydrogen atoms of the methyl groups of pyruvate with protons of the solvent.[10]
Evolutionary significance
History
Arguments have been made for both the convergent and divergent evolution of α/β-barrel structured enzymes such as KDPG Aldolase, triosephosphate isomerase, and the A-domain of pyruvate kinase.
Convergent evolution can lead to geometrically similar active sites while each enzyme has a distinct backbone conformation. Convergence to a common backbone structure, as is the case here however, has not been observed, although it is argues that it might be possible for a symmetrically repetitive structure as the one observed here.[11] The similarity in the folding of the three enzymes and the exceptional symmetry commonly suggests divergent evolution from a common ancestor. The functional similarity of the enzymes remains the strongest argument for divergent evolution.[11] All three enzymes activate a C–H bond adjacent to a carbonyl group. The active sites are located at the carboxylic ends of the β strands. Such congruence is in favor of divergent evolution.
Should the divergent evolution hypothesis prevail, this would suggest the existence of a class of enzymes with unrelated amino acid sequences yet analogous symmetrical structure and folding.[11]
Directed Evolution
KDPG aldolase has limited utility due to its high specificity for its natural substrates in the cleavage of KDPG and the reverse addition of D-glyceraldehyde-3-phosphate and pyruvate.[12] In vitro evolution has allowed KDPG aldolase to be converted into a more efficient aldolase with altered substrate specificity and stereoselectivity thereby improving its utility in asymmetric synthesis.[13] Rather than modifying the recognition site, the substrate is modified by moving the active site lysine from one β strand to a neighboring one.[13][14] The evolved aldolase is capable of accepting both D- and L-glyceraldehyde in their non-phosphorylated form.[15]
References
- Peekhaus N, Conway T (July 1998). "What's for dinner?: Entner–Doudoroff metabolism in Escherichia coli". J. Bacteriol. 180 (14): 3495–502. PMC 107313. PMID 9657988.
- Mavridis IM, Tulinsky A (October 1976). "The folding and quaternary structure of trimeric 2-keto-3-deoxy-6-phosphogluconic aldolase at 3.5-A resolution". Biochemistry. 15 (20): 4410–7. doi:10.1021/bi00665a010. PMID 974067.
- Mavridis IM, Hatada MH, Tulinsky A, Lebioda L (December 1982). "Structure of 2-keto-3-deoxy-6-phosphogluconate aldolase at 2 . 8 A resolution". J. Mol. Biol. 162 (2): 419–44. doi:10.1016/0022-2836(82)90536-8. PMID 7161801.
- Suzuki N, Wood WA (April 1980). "Complete primary structure of 2-keto-3-deoxy-6-phosphogluconate aldolase". J. Biol. Chem. 255 (8): 3427–35. PMID 6988426.
- Richardson JS (September 1979). "The singly-wound parallel beta barrel: a proposed structure for 2-keto-3-deoxy-6-phosphogluconate aldolase". Biochem. Biophys. Res. Commun. 90 (1): 285–90. doi:10.1016/0006-291X(79)91622-X. PMID 496979.
- Bell BJ, Watanabe L, Rios-Steiner JL, Tulinsky A, Lebioda L, Arni RK (August 2003). "Structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida". Acta Crystallogr. D. 59 (Pt 8): 1454–8. doi:10.1107/S0907444903013192. PMID 12876349.
- Fullerton SW, Griffiths JS, Merkel AB, et al. (May 2006). "Mechanism of the Class I KDPG aldolase". Bioorg. Med. Chem. 14 (9): 3002–10. doi:10.1016/j.bmc.2005.12.022. PMC 3315828. PMID 16403639.
- Möhler H, Decker K, Wood WA (July 1972). "Structure of 2-keto-3-deoxy-6-phosphogluconate aldolase. IV. Structural features revealed by treatment with urea and Ellman's reagent". Arch. Biochem. Biophys. 151 (1): 251–60. doi:10.1016/0003-9861(72)90495-x. PMID 5044518.
- Allard J, Grochulski P, Sygusch J (March 2001). "Covalent intermediate trapped in 2-keto-3-deoxy-6- phosphogluconate (KDPG) aldolase structure at 1.95-A resolution". Proc. Natl. Acad. Sci. U.S.A. 98 (7): 3679–84. doi:10.1073/pnas.071380898. PMC 31111. PMID 11274385.
- Meloche HP, Glusker JP (July 1973). "Aldolase catalysis: single base-mediated proton activation". Science. 181 (4097): 350–2. doi:10.1126/science.181.4097.350. PMID 4719907.
- Lebioda L, Hatada MH, Tulinsky A, Mavridis IM (December 1982). "Comparison of the folding of 2-keto-3-deoxy-6-phosphogluconate aldolase, triosephosphate isomerase and pyruvate kinase. Implications in molecular evolution". J. Mol. Biol. 162 (2): 445–58. doi:10.1016/0022-2836(82)90537-X. PMID 7161802.
- Farinas ET, Bulter T, Arnold FH (December 2001). "Directed enzyme evolution". Curr. Opin. Biotechnol. 12 (6): 545–51. doi:10.1016/S0958-1669(01)00261-0. PMID 11849936.
- Fong S, Machajewski TD, Mak CC, Wong C (November 2000). "Directed evolution of D-2-keto-3-deoxy-6-phosphogluconate aldolase to new variants for the efficient synthesis of D- and L-sugars". Chem. Biol. 7 (11): 873–83. doi:10.1016/S1074-5521(00)00035-1. PMID 11094340.
- Wymer N, Buchanan LV, Henderson D, et al. (January 2001). "Directed evolution of a new catalytic site in 2-keto-3-deoxy-6-phosphogluconate aldolase from Escherichia coli". Structure. 9 (1): 1–9. doi:10.1016/S0969-2126(00)00555-4. PMID 11342129.
- Huisman GW, Gray D (August 2002). "Towards novel processes for the fine-chemical and pharmaceutical industries". Curr. Opin. Biotechnol. 13 (4): 352–8. doi:10.1016/S0958-1669(02)00335-X. PMID 12323358.
Further reading
- Meloche HP, Wood WA (1964). "Crystallization and characteristics of 2-keto-3-deoxy-6-phosphogluconic aldolase". J. Biol. Chem. 239: 3515–3518. PMID 14245411.
External links
