Realm (virology)
In virology, a realm is the highest taxonomic rank used for viruses, established by the International Committee on Taxonomy of Viruses (ICTV), which oversees virus taxonomy. Four realms are recognized: Duplodnaviria, Monodnaviria, Riboviria, and Varidnaviria. While the rank of realm corresponds to the rank of domain used for cellular life, it chiefly differs in that viruses in a realm do not necessarily share ancestry via common descent, but are rather grouped together based on specific traits that are highly conserved over long periods of time. Likewise, virus realms generally do not have any genetic relation to each other. As such, each realm represents at least one instance of viruses coming into existence.
Naming
The names of realms consist of a descriptive first part and the suffix -viria, which is the suffix used for virus realms.[1] The first part of Duplodnaviria means "double DNA", referring to dsDNA viruses,[2] the first part of Monodnaviria means "single DNA", referring to ssDNA viruses,[3] the first part of Riboviria is taken from ribonucleic acid (RNA),[4] and the first part of Varidnaviria means "various DNA".[5] For viroids, the suffix is designated as -viroidia, and for satellites, the suffix is -satellitia,[1] but as of 2019 neither viroid nor satellite realms have been designated.[6]
Realms
Duplodnaviria
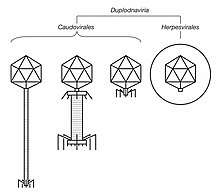
Duplodnaviria contains double-stranded DNA (dsDNA) viruses that encode the HK97 major capsid protein (MCP). Viruses in the realm also share a number of other characteristics involving the capsid and capsid assembly, including an icosahedral capsid shape and a terminase enzyme that packages viral DNA into the capsid during assembly. Two groups of viruses are included in the realm: tailed bacteriophages, assigned to the order Caudovirales, and herpesviruses, assigned to the order Herpesvirales.[2]
Monodnaviria
Monodnaviria contains single-stranded DNA (ssDNA) viruses that encode an endonuclease of the HUH superfamily that initiates rolling circle replication and all other viruses descended from such viruses. The prototypical members of the realm are called CRESS-DNA viruses and have circular ssDNA genomes. ssDNA viruses with linear genomes are descended from them, and in turn some dsDNA viruses with circular genomes are descended from the linear ssDNA viruses.[3]
Riboviria
Riboviria contains all RNA viruses, assigned to the kingdom Orthornavirae, and all reverse transcribing viruses, assigned to the kingdom Pararnavirae. RNA viruses are marked by their RNA-dependent RNA polymerase (RdRp), which transcribes viral mRNA and replicates the genome, whereas reverse transcribing viruses encode and replicate via reverse transcriptase. Most eukaryotic viruses, including most human, animal, and plant viruses, belong to Riboviria.[4]
Varidnaviria
Varidnaviria contains DNA viruses that encode MCPs that have a jelly roll fold folded structure in which the jelly roll fold is perpendicular to the surface of the capsid. Many members also share many other characteristics, including a minor capsid protein with a single jelly roll fold, an ATPase that packages the genome, and a common DNA polymerase. Two kingdoms are recognized: Helvetiavirae, whose members have MCPs with a single jelly roll fold, and Bamfordvirae, whose members have MCPs with two jelly roll folds.[5]
Origins
In general, virus realms have no genetic relation to each other based on common descent, in contrast to the three domains of cellular life—Archaea, Bacteria, and Eukarya—which share a common ancestor. Likewise, viruses within each realm are not necessarily descended from a common ancestor since realms group viruses together based highly conserved traits, not common ancestry, which is used as the basis for the taxonomy of cellular life. As such, each virus realm is considered to represent at least one instance of viruses coming into existence.[7] By realm:
- Duplodnaviria is either monophyletic or polyphyletic and may predate the last universal common ancestor (LUCA) of cellular life. The exact origin of the realm is not known, but the HK97 MCP encoded by all members is, outside the realm, only found in encapsulins, a type of nanocompartment found in bacteria, although the relation between Duplodnaviria and encapsulins is fully understood.[2][8][9]
- Monodnaviria is polyphyletic, having emerged multiple times from bacterial and archaeal circular plasmids, which are extra-chromosomal DNA molecules that live inside of bacteria and archaea and which self-replicate.[3][10]
- Riboviria is monophyletic or polyphyletic with two origins. The reverse transcriptase of kingdom Pararnavirae evolved from a retrotransposon, a type of self-replicating DNA molecule that replicates via reverse transcription, on a single occasion. The origin of the RdRp of Orthornavirae is less certain, but it is likewise believed to originate from a retroelement such as a retrotransposon or a group II intron that encodes reverse transcriptase.[4][11]
- Varidnaviria is either monophyletic or polyphyletic and may predate the LUCA. The kingdom Bamfordvirae is derived from the other kingdom Helvetiavirae via fusion of two MCPs to have an MCP with two jelly roll folds instead of one. The single jelly roll (SJR) fold MCPs of Helvetiavirae show a relation to a group of proteins that contain SJR folds, including the Cupin superfamily and nucleoplasmins.[5][8][9]
While the realms generally have no genetic relation to each other, there are some exceptions:
- Viruses in the family Podoviridae in Duplodnaviria encode a DNA polymerase that is related to the DNA polymerases encoded by many members of Varidnaviria.[12]
- Eukaryotic viruses in the kingdom Shotokuvirae in Monodnaviria were created on multiple occasions by recombination events that combined the DNA of ancestral plasmids with complementary DNA (cDNA) of positive sense RNA viruses in Riboviria, by which ssDNA viruses in Shotokuvirae obtained capsid proteins from RNA viruses.[3][8]
- The family Bidnaviridae in Monodnaviria was created via integration of a parvovirus (of Monodnaviria) genome into a polinton, a virus-like self-replicating DNA molecule, which are related to viruses in Varidnaviria. Furthermore, bidnaviruses encode a receptor-binding protein inherited from reoviruses in the realm Riboviria.[13]
Subrealm
In virology, the second highest taxonomy rank established by the ICTV is subrealm, which is the rank below realm. Subrealms of viruses use the suffix -vira, viroid subrealms use the suffix -viroida, and satellites use the suffix -satellitida. The rank below subrealm is kingdom. As of 2019, no taxa are described at the rank of subrealm.[1][6]
History
In the 21st century, the identification of many new viruses via metagenomics and comparison of highly conserved traits in viruses has enabled deep evolutionary relationships between them to be discovered. This led to the desire to create higher-level taxonomy for viruses. In two votes in 2018 and 2019, the ICTV agreed to adopt a 15-rank classification system for viruses, ranging from realm to species, replacing the prior system that ranged from order to species.[7] As part of this reform, Riboviria was established as the first realm in 2018,[14] and the other three realms were established a year later.[15][16][17]
See also
References
- "ICTV Code The International Code of Virus Classification and Nomenclature". International Committee on Taxonomy of Viruses (ICTV). October 2018. Retrieved 18 March 2020.
- Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini M, Kuhn JH (18 October 2019). "Create a megataxonomic framework, filling all principal/primary taxonomic ranks, for dsDNA viruses encoding HK97-type major capsid proteins" (docx). International Committee on Taxonomy of Viruses. Retrieved 13 August 2020.
- Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini M, Kuhn JH (18 October 2019). "Create a megataxonomic framework, filling all principal taxonomic ranks, for ssDNA viruses" (docx). International Committee on Taxonomy of Viruses. Retrieved 13 August 2020.
- Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini M, Kuhn JH (18 October 2019). "Create a megataxonomic framework, filling all principal taxonomic ranks, for realm Riboviria" (docx). International Committee on Taxonomy of Viruses (ICTV). Retrieved 13 August 2020.
- Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini M, Kuhn JH (18 October 2019). "Create a megataxonomic framework, filling all principal taxonomic ranks, for DNA viruses encoding vertical jelly roll-type major capsid proteins" (docx). International Committee on Taxonomy of Viruses. Retrieved 13 August 2020.
- "Virus Taxonomy: 2019 Release". International Committee on Taxonomy of Viruses. International Committee on Taxonomy of Viruses. Retrieved 25 April 2020.
- International Committee on Taxonomy of Viruses Executive Committee (May 2020). "The New Scope of Virus Taxonomy: Partitioning the Virosphere Into 15 Hierarchical Ranks". Nat Microbiol. 5 (5): 668–674. doi:10.1038/s41564-020-0709-x. PMC 7186216. PMID 32341570. Retrieved 13 August 2020.
- Krupovic M, Koonin EV (21 March 2017). "Multiple origins of viral capsid proteins from cellular ancestors". Proc Natl Acad Sci U S A. 114 (12): E2401–E2410. doi:10.1073/pnas.1621061114. PMC 5373398. PMID 28265094.
- Krupovic, M; Dolja, VV; Koonin, EV (14 July 2020). "The LUCA and its complex virome". Nat Rev Microbiol. doi:10.1038/s41579-020-0408-x. PMID 32665595.
- Kazlauskas D, Varsani A, Koonin EV, Krupovic M (31 July 2019). "Multiple Origins of Prokaryotic and Eukaryotic Single-Stranded DNA Viruses From Bacterial and Archaeal Plasmids". Nat Commun. 10 (1): 3425. doi:10.1038/s41467-019-11433-0. PMC 6668415. PMID 31366885. Retrieved 13 August 2020.
- Wolf YI, Kazlauskas D, Iranzo J, Lucia-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Kooning EV (27 November 2018). "Origins and Evolution of the Global RNA Virome". mBio. 9 (6): e02329-18. doi:10.1128/mBio.02329-18. PMC 6282212. PMID 30482837.
- Krupovic M, Koonin EV (February 2015). "Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution". Nat Rev Microbiol. 13 (2): 105–115. doi:10.1038/nrmicro3389. PMC 5898198. PMID 25534808. Retrieved 13 August 2020.
- Krupvoic M, Koonin EV (18 June 2014). "Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses". Sci Rep. 4: 5347. doi:10.1038/srep05347. PMC 4061559. PMID 24939392. Retrieved 13 August 2020.
- "ICTV Taxonomy history: Riboviria". International Committee on Taxonomy of Viruses. International Committee on Taxonomy of Viruses. February 2019. Retrieved 13 August 2020.
- "ICTV Taxonomy history: Duplodnaviria". International Committee on Taxonomy of Viruses. International Committee on Taxonomy of Viruses. March 2020. Retrieved 13 August 2020.
- "ICTV Taxonomy history: Monodnaviria". International Committee on Taxonomy of Viruses. International Committee on Taxonomy of Viruses. March 2020. Retrieved 13 August 2020.
- "ICTV Taxonomy history: Varidnaviria". International Committee on Taxonomy of Viruses. International Committee on Taxonomy of Viruses. March 2020. Retrieved 13 August 2020.
Further reading
- Ward, C. W. (1993). "Progress towards a higher taxonomy of viruses". Research in Virology. 144 (6): 419–53. doi:10.1016/S0923-2516(06)80059-2. PMC 7135741. PMID 8140287.