SmY RNA
SmY ribonucleic acids (SmY RNAs) are a family of small nuclear RNAs found in some species of nematode worms. They are thought to be involved in mRNA trans-splicing.
| SmY spliceosomal RNA | |
|---|---|
 Consensus secondary structure of SmY RNAs.
A count of base pair substitutions observed in 68 structurally aligned sequences in the Rfam seed alignment is shown for each base pair, illustrating the extensive support for this predicted structure.[1] | |
| Identifiers | |
| Symbol | SmY |
| Rfam | RF01844 |
| Other data | |
| RNA type | snRNA, splicing |
| Domain(s) | Chromadorea |
| PDB structures | PDBe |
SmY RNAs are about 70–90 nucleotides long and share a common secondary structure, with two stem-loops flanking a consensus binding site for Sm protein.[2][3] Sm protein is a shared component of spliceosomal snRNPs.
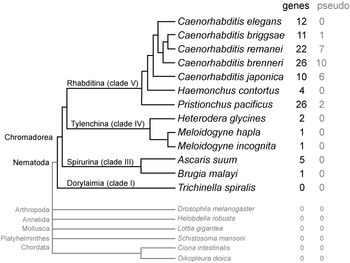
SmY RNAs have been found in nematodes of class Chromadorea, which includes the most commonly studied nematodes (such as Caenorhabditis, Pristionchus, and Ascaris), but not in the more distantly related Trichinella spiralis in class Dorylaimia. The number of SmY genes in each species varies, with most Caenorhabditis and Pristionchus species having 10–26 related paralogous copies, while other nematodes have 1–5.[1]
Discovery
The first SmY RNA was discovered in 1996 in purified Ascaris lumbricoides spliceosome preparations, as was another called SmX RNA that is not detectably homologous to SmY.[2] Twelve SmY homologs were identified computationally in Caenorhabditis elegans, and ten in Caenorhabditis briggsae.[3] Several transcripts from these SmY genes were cloned and sequenced in a systematic survey of small non-coding RNA transcripts in C. elegans.[4]
A systematic survey of 2,2,7-trimethylguanosine (TMG) 5' capped transcripts in C.elegans using anti TMG antibodies identified two TMG capped SmY transcripts.[5] Sequence analysis of the potential Sm binding sites in these transcripts indicated the SmY, U5 snRNA, U3 snoRNA and the spliced leader RNAs transcripts (SL1 and SL2) all contain a very similar consensus SM binding sequence ( AAU4–5GGA). The predicted SM binding sites identified in the U1, U2 and U4 snRNA transcripts varied from this consensus.[5]
Function
In C. elegans, SmY RNAs copurify with spliceosome and with Sm, SL75p, and SL26p proteins, while the better-characterized C. elegans SL1 trans-splicing snRNA copurifies in a complex with Sm, SL75p, and SL21p (a paralog of SL26p).[2][3] Loss of function of either SL21p or SL26p individually causes only a weak cold-sensitive phenotype, whereas knockdown of both is lethal, as is a SL75p knockdown. Based on these results, the SmY RNAs are believed to have a function in trans-splicing.
References
- Jones TA, Otto W, Marz M, Eddy SR, Stadler PF (2009). "A survey of nematode SmY RNAs". RNA Biol. 6 (1): 5–8. doi:10.4161/rna.6.1.7634. PMID 19106623.
- Maroney PA, Yu YT, Jankowska M, Nilsen TW (August 1996). "Direct analysis of nematode cis- and trans-spliceosomes: a functional role for U5 snRNA in spliced leader addition trans-splicing and the identification of novel Sm snRNPs". RNA. 2 (8): 735–745. PMC 1369411. PMID 8752084.
- MacMorris M, Kumar M, Lasda E, Larsen A, Kraemer B, Blumenthal T (April 2007). "A novel family of C. elegans snRNPs contains proteins associated with trans-splicing". RNA. 13 (4): 511–520. doi:10.1261/rna.426707. PMC 1831854. PMID 17283210.
- Deng W, Zhu X, Skogerbø G, et al. (January 2006). "Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression". Genome Research. 16 (1): 20–29. doi:10.1101/gr.4139206. PMC 1356125. PMID 16344563.
- Jia, D.; Cai, L.; He, H.; Skogerbø, G.; Li, T.; Aftab, N.; Chen, R. (September 2007). "Systematic identification of non-coding RNA 2,2,7-trimethylguanosine cap structures in Caenorhabditis elegans". BMC Molecular Biology. 8: 86. doi:10.1186/1471-2199-8-86. PMC 2200864. PMID 17903271.