U2 spliceosomal RNA
U2 spliceosomal snRNAs are a species of small nuclear RNA (snRNA) molecules found in the major spliceosomal (Sm) machinery of virtually all-eukaryotic organisms. In vivo, U2 snRNA along with its associated polypeptides assemble to produce the U2 small nuclear ribonucleoprotein (snRNP), an essential component of the major spliceosomal complex.[1] The major spliceosomal-splicing pathway is occasionally referred to as U2 dependent, based on a class of Sm intron—found in mRNA primary transcripts—that are recognized exclusively by the U2 snRNP during early stages of spliceosomal assembly.[2] In addition to U2 dependent intron recognition, U2 snRNA has been theorized to serve a catalytic role in the chemistry of pre-RNA splicing as well.[3][4] Similar to ribosomal RNAs (rRNAs), Sm snRNAs must mediate both RNA:RNA and RNA:protein contacts and hence have evolved specialized, highly conserved, primary and secondary structural elements to facilitate these types of interactions.[5][6]
| U2 spliceosomal RNA | |
|---|---|
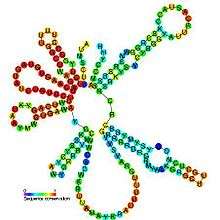 Predicted secondary structure and sequence conservation of U2 | |
| Identifiers | |
| Symbol | U2 |
| Rfam | RF00004 |
| Other data | |
| RNA type | Gene; snRNA; splicing |
| Domain(s) | Eukaryota |
| GO | 0000370 0045131 0005686 |
| SO | 0000392 |
| PDB structures | PDBe |
Shortly after the discovery that mRNA primary transcripts contain long, non-coding intervening sequences (introns) by Sharp and Roberts,[7][8] Joan Steitz began work to characterize the biochemical mechanism of intron excision.[9] The curious observation that a sequence found in the 5´ region of the U1 snRNA exhibited extensive base pairing complementarity with conserved sequences across 5´ splice junctions in hnRNA transcripts prompted speculation that certain snRNAs may be involved in recognizing splice site boundaries through RNA:RNA contacts.[9] Only recently have atomic crystal structures revealed demonstrably that the original conjecture was indeed correct, even if the complexity of these interactions were not fully realized at the time.[5][6][10]
U2 snRNA Recognition Elements
In Saccharomyces cerevisiae the U2 snRNA is associated with 18 polypeptides, seven of which are structural proteins common to all Sm class snRNPs.[11] These non-specific structural proteins associate with Sm snRNAs through a highly conserved recognition sequence (AUnG,n = 4-6) located within the RNA called Sm-binding sites.[12] Two other proteins, A´ and B´´, are U2-specific and require structural elements unique to U2 snRNA—specifically two 3´ stem loops—for snRNP assembly.[11] The three-subunit SF3a and six-subunit SF3b protein complexes also associate with the U2 snRNA[13].
U2 snRNA is implicated in intron recognition through a 7-12 nucleotide sequence between 18-40 nucleotides upstream of the 3´ splice site known as the branch point sequence (BPS).[1][2] In yeast, the consensus BPS is 7 nucleotide residues in length and the complementary recognition sequence within the U2 snRNA is 6 nucleotides. Duplex formation between these two sequences results in bulging of a conserved adenosine residue at position 5 of the BPS. The bulged adenosine residue adopts a C3´-endo conformation[14] that with the help of splicing factors Cwc25, Yju2 and Isy1 aligns a 2´ OH for an inline attack of a phosphorus atom at the 5´ splice site[15]. Nucleophilic attack initiates the first of two successive transesterification reactions that splices out the intron—through an unusual 2´-5´-3´ linked lariat intermediate—where the second transesterification involves ligation of the two flanking exons.
Primary and Secondary Structure
Although the sequence length of U2 snRNAs can vary by up to an order of magnitude across all eukaryotic organisms, all U2 snRNAs contain many phylogenetically constant regions particularly within the first 80 nucleotides downstream of the 5´ end where 85% of the positions are conserved.[16] Moreover, several secondary structural elements are also conserved including stem loops I, II, III, IV, and some of the single stranded regions linking these domains.[16][17] Stem loop II in yeast U2 snRNA, contains an unusual sheared GA base pair leading into a characteristic U-turn loop motif that shares a geometric conformation similar to that of tRNA anti-codon loops.[5] All U2 snRNAs possess a terminal stem loop (IV) with a 10-16 base pair helix and a conserved 11 nucleotide loop with the consensus sequence 5´-UYGCANUURYN-3´.[16]
U2 snRNAs are the most extensively modified of all the small nuclear RNAs.[18] While the exact locations of these post-transcriptional modifications can vary from organism to organism, emerging evidence suggests there is a strong correlation between U2 snRNA modification and biological function.[18] Modifications include the conversion of some uridine residues to pseudouridine, 2´-O-methylation, nucleobase methylation, and conversion of 5´-monomethylated guanosine cap to a 2,2,7-trimethylated guanosine cap.[18] Many of these modifications reside in a 27-nucleotide region on the 5´ end of the molecule.[18]
Conformational Dynamics
The spliceosome is a dynamic molecular machine that undergoes several conformational rearrangements throughout assembly and splicing. Although many of the biochemical details surrounding spliceosomal rearrangements remains unclear, recent studies have visualized the formation of a critical folding complex between U2 and U6 snRNAs immediately proceeding the first step of the splicing reaction.[19][6] This folding event facilitates the formation of a four-helix junction, which is believed to provide scaffolding for the critical components of the active site including aligning the 5´ splice site with the branch point adenosine for inline attack by the 2´ OH and coordinating two Mg2+ ions to stabilize negative charge formation in the proceeding steps.[19]
Evolutionary Origins
A notable characteristic of the U2-U6 fold is its structural similarity to that of domain V in self-splicing group II introns.[6][3] The AGC triad found in U6 snRNA is conserved in group II introns and has been found to favor the same tertiary stacking interactions as well.[6] The formation of a GU wobble pair early in the U2-U6 folding event is also observed in the formation of the catalytic core of group II introns.[19] Finally, it is likely the spliceosome utilizes the same two-metal ion mechanism as group II introns given the structural conservation of metal binding sites found within the U2-U6 fold.[3] The extent of both secondary and tertiary structure conservation between group II introns and the U2-U6 fold in the active site of the spliceosome strongly suggests both group II introns and the spliceosome share a common evolutionary origin.
References
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002). Molecular Biology of the Cell (4th ed.). Garland Science. ISBN 978-0815332183.
- Nelson DL, Cox MM, Lehninger AL (2013). Lehninger principles of biochemistry (6th ed.). New York: W.H. Freeman and Company. ISBN 9781429234146. OCLC 824794893.
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA (November 2013). "RNA catalyses nuclear pre-mRNA splicing". Nature. 503 (7475): 229–34. Bibcode:2013Natur.503..229F. doi:10.1038/nature12734. PMC 4666680. PMID 24196718.
- Shi Y (August 2017). "The Spliceosome: A Protein-Directed Metalloribozyme". Journal of Molecular Biology. 429 (17): 2640–2653. doi:10.1016/j.jmb.2017.07.010. PMID 28733144.
- Stallings, Sarah C; Moore, Peter B (1997). "The structure of an essential splicing element: stem loop IIa from yeast U2 snRNA". Structure. 5 (9): 1173–1185. doi:10.1016/s0969-2126(97)00268-2. ISSN 0969-2126. PMID 9331416.
- Burke JE, Sashital DG, Zuo X, Wang YX, Butcher SE (April 2012). "Structure of the yeast U2/U6 snRNA complex". RNA. 18 (4): 673–83. doi:10.1261/rna.031138.111. PMC 3312555. PMID 22328579.
- Berget SM, Moore C, Sharp PA (August 1977). "Spliced segments at the 5' terminus of adenovirus 2 late mRNA". Proceedings of the National Academy of Sciences of the United States of America. 74 (8): 3171–5. Bibcode:1977PNAS...74.3171B. doi:10.1073/pnas.74.8.3171. PMC 431482. PMID 269380.
- Chow LT, Gelinas RE, Broker TR, Roberts RJ (September 1977). "An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA". Cell. 12 (1): 1–8. doi:10.1016/0092-8674(77)90180-5. PMID 902310.
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA (January 1980). "Are snRNPs involved in splicing?". Nature. 283 (5743): 220–4. doi:10.1038/283220a0. PMID 7350545.
- Perriman R, Ares M (May 2010). "Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing". Molecular Cell. 38 (3): 416–27. doi:10.1016/j.molcel.2010.02.036. PMC 2872779. PMID 20471947.
- Pan ZQ, Prives C (December 1989). "U2 snRNA sequences that bind U2-specific proteins are dispensable for the function of U2 snRNP in splicing". Genes & Development. 3 (12A): 1887–98. doi:10.1101/gad.3.12a.1887. PMID 2559872.
- Mattaj IW, Habets WJ, van Venrooij WJ (May 1986). "Monospecific antibodies reveal details of U2 snRNP structure and interaction between U1 and U2 snRNPs". The EMBO Journal. 5 (5): 997–1002. doi:10.1002/j.1460-2075.1986.tb04314.x. PMC 1166893. PMID 2941274.
- Dziembowski, Andrzej; Ventura, Ana-Paula; Rutz, Berthold; Caspary, Friederike; Faux, Céline; Halgand, Frédéric; Laprévote, Olivier; Séraphin, Bertrand (2004-12-08). "Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing". The EMBO Journal. 23 (24): 4847–4856. doi:10.1038/sj.emboj.7600482. ISSN 0261-4189. PMC 535094. PMID 15565172.
- Berglund JA, Rosbash M, Schultz SC (May 2001). "Crystal structure of a model branchpoint-U2 snRNA duplex containing bulged adenosines". RNA. 7 (5): 682–91. doi:10.1017/S1355838201002187. PMC 1370120. PMID 11350032.
- Galej, Wojciech P.; Wilkinson, Max E.; Fica, Sebastian M.; Oubridge, Chris; Newman, Andrew J.; Nagai, Kiyoshi (8 September 2016). "Cryo-EM structure of the spliceosome immediately after branching". Nature. 537 (7619): 197–201. Bibcode:2016Natur.537..197G. doi:10.1038/nature19316. ISSN 1476-4687. PMC 5156311. PMID 27459055.
- Guthrie C, Patterson B (1988). "Spliceosomal snRNAs". Annual Review of Genetics. 22: 387–419. doi:10.1146/annurev.ge.22.120188.002131. PMID 2977088.
- Krämer A (August 1987). "Analysis of RNase-A-resistant regions of adenovirus 2 major late precursor-mRNA in splicing extracts reveals an ordered interaction of nuclear components with the substrate RNA". Journal of Molecular Biology. 196 (3): 559–73. doi:10.1016/0022-2836(87)90032-5. PMID 3681967.
- Yu YT, Shu MD, Steitz JA (October 1998). "Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing". The EMBO Journal. 17 (19): 5783–95. doi:10.1093/emboj/17.19.5783. PMC 1170906. PMID 9755178.
- Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE (December 2004). "U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction". Nature Structural & Molecular Biology. 11 (12): 1237–42. doi:10.1038/nsmb863. PMID 15543154.
Further reading
- Newby MI, Greenbaum NL (June 2001). "A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture". RNA. 7 (6): 833–45. doi:10.1017/S1355838201002308. PMC 1370140. PMID 11424937.
- Berglund JA, Rosbash M, Schultz SC (May 2001). "Crystal structure of a model branchpoint-U2 snRNA duplex containing bulged adenosines". RNA. 7 (5): 682–91. doi:10.1017/S1355838201002187. PMC 1370120. PMID 11350032.