Pseudouridine
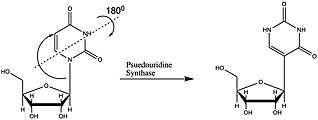
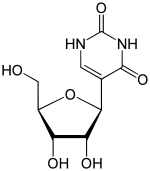
Pseudouridine (abbreviated by the Greek letter psi- Ψ or the letter Q)[1] is an isomer of the nucleoside uridine in which the uracil is attached via a carbon-carbon instead of a nitrogen-carbon glycosidic bond. (In this configuration, uracil is sometimes referred to as 'pseudouracil'.) Pseudouridine is the most abundant RNA modification in cellular RNA. After transcription and following synthesis, RNA can be modified with over 100 chemically distinct modifications. These can potentially regulate RNA expression post-transcriptionally, in addition to the four standard nucleotides and play a variety of roles in the cell including translation, localization and stabilization of RNA. Pseudouridine, being one of them, is the C5-glycoside isomer of uridine that contains a C-C bond between C1 of the ribose sugar and C5 of uracil, rather than usual C1-N1 bond found in uridine. The C-C bond gives it more rotational freedom and conformational flexibility.[2] In addition, pseudouridine has an extra hydrogen bond donor at the N1 position. Also known as 5-ribosyluracil, pseudouridine is a ubiquitous yet enigmatic constituent of structural RNAs (transfer, ribosomal, small nuclear (snRNA) and small nucleolar). Recently it has also been discovered in coding RNA. Being the most abundant, it is found in all 3 phylogenetic domains of life and was the first to be discovered. This nucleotide is considered to be the “fifth nucleotide”. It accounts for 4% of the nucleotides in the yeast tRNA. This base modification is able to stabilize RNA and improve base-stacking by forming additional hydrogen bonds with water through its extra imino group. There are 11 pseudouridines in the Escherichia coli rRNA, 30 in yeast cytoplasmic rRNA and a single modification in mitochondrial 21S rRNA and about 100 pseudouridines in human rRNA indicating that the extent of pseudouridylation increases with the complexity of an organism. Pseudouridine in rRNA and tRNA has been shown to fine-tune and stabilize the regional structure and help maintain their functions in mRNA decoding, ribosome assembly, processing and translation.[2][3][4] Pseudouridine in snRNA has been shown to enhance spliceosomal RNA-pre-mRNA interaction to facilitate splicing regulation.[5]
 | |
| Names | |
|---|---|
| IUPAC name
5-[(2S,3R,4S,5R)-3,4-Dihydroxy-5-(hydroxymethyl)-oxolan-2-yl]-1H-pyrimidine-2,4-dione | |
| Preferred IUPAC name
5-(β-D-ribofuranosyl)pyrimidine-2,4(1H,3H)-dione | |
| Other names
psi-Uridine, 5-Ribosyluracil, beta-D-Pseudouridine, 5-(beta-D-Ribofuranosyl)uracil | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChemSpider | |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C9H12N2O6 | |
| Molar mass | 244.20 g/mol |
| Appearance | White granular powder |
| Highly soluble in water. | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Effects and modification on different RNA
tRNA
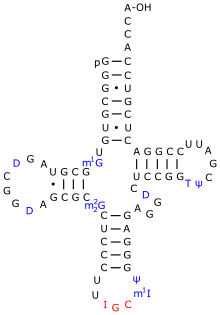
Pseudouridine = Ψ
Ψ is ubiquitous in this class of RNAs and facilitates common tRNA structural motifs. One such structural motif is the TΨC stem loop which incorporates Ψ55. Ψ is commonly found in the D stem and anticodon stem and loop of tRNAs from each domain. In each structural motif the unique physicochemical properties of Ψ stabilize structures that would not be possible with the standard U.[2]
During translation Ψ modulates interactions of tRNA molecules with rRNAs and mRNAs. Ψ and other modified nucleotides affect the local structure of the tRNA domains they are found in without impacting the overall fold of the RNA. In the anticodon stem-loop (ASL) Ψ seems critical for proper binding of tRNAs to the ribosome. Ψ stabilizes the dynamic structure of the ASL and promotes stronger binding to the 30S ribosome. The stabilized conformation of the ASL helps maintain correct anticodon-codon pairings during translation. This stability may increase translational accuracy by decreasing the rate of peptide bond formation and allowing for more time for incorrect codon-anticodon pairs to be rejected. Despite Ψ’s role in local structure stabilization, pseudouridylation of tRNA is not essential for cell viability and is not usually required for aminoacylation.[2]
mRNA
Ψ is also found in mRNAs which are the template for protein synthesis. Ψ residues in mRNA can affect the coding specificity of stop codons UAA, UGA, and UAG. In these stop codons both a U→Ψ modification and a U→C mutation both promote nonsense suppression.[6]
rRNA
Ψ is found in the large and small ribosomal subunits of all domains of life and their organelles. In the ribosome Ψ residues cluster in domains II, IV, and V and stabilize RNA-RNA and/or RNA-protein interactions. The stability afforded by Ψ may assist rRNA folding and ribosome assembly. Ψ may also influence the stability of local structures which impact the speed and accuracy of decoding and proofreading during translation.[2]
snRNA
Ψ is found in the major spliceosomal snRNAs of eukaryotes. Ψ residues in snRNA are often phylogenetically conserved, but have some variations across taxa and organisms. The Ψ residues in snRNAs are normally located in regions that participate in RNA-RNA and/or RNA-protein interactions involved in the assembly and function of the spliceosome. Ψ residues in snRNAS contribute to the proper folding and assembly of the spliceosome which is essential for pre-mRNA processing.[2]
Pseudouridine synthase proteins
Pseudouridine are RNA modifications that are done post-transcription, so after the RNA is formed. The proteins that do this modification are called Pseudouridine Synthases or PUS. PUS proteins are found in all kingdoms of life. Most research has been done on how PUS protein modify tRNA, so mechanism involving snRNA, and mRNA are not clearly defined. PUS proteins can vary on RNA specificity, structure , and isomerization mechanisms. The different structures of PUS are divided into 4 families. The families share the active sequence and important structural motifs.[1]
TruA
TruA domain modifies a variety of different places in tRNA, snRNA, and mRNA. The mechanism of isomerization of uridine is still being talked about in this family.[4][7]
PUS 1 is located in the nucleus and modifies tRNA at different locations, U44 of U2 snRNA, and U28 of U6 snRNA. Studies found that PUS 1 expression increased during environmental stress and is important for regulating the splicing of RNA. Also that PUS 1 is necessary for taking the tRNA made in the nucleus and sending them to the cytoplasm.[4]
PUS 2 is very similar to PUS 1, but located in the mitochondria and only modifies U27 and U28 of mito-tRNA. This protein modifies the mitochondrial tRNA, which have a lesser amount of pseudouridine modifications compared to other tRNA. Unlike most mitochondria located protein, PUS 2 has not been found to have a mitochondrial targeting signal or MTS.[4]
PUS 3 is a homolog to PUS 1, but modifies different places of the tRNA (U38/39) in the cytoplasm and mitochondrial. This protein is the most conserved of the TruA family. A decrease in modifications made by PUS 3 was found when the tRNA structure of improperly folded. Along with tRNA the protein targets ncRNA and mRNA, further research is still needed as to the importance of this modification. PUS 3 along with PUS 1 modify steroid activator receptor in humans.[4]
TruB
The TruB family only contains PUS 4 located in the mitochondrial and nucleus. PUS 4 modification is heavily conserved located in the U55 in the elbow of the tRNA. The human form of PUS 4 is actually missing a binding domain called PUA or pseudouridine synthase and archaeosine trans-glycosylase. PUS 4 has a sequence specificity for T-loop part of the tRNA. Preliminary data of PUS4 modifying mRNA, but more research is needed to confirm. Also binds to a specific Brome Mosaic Virus, which is a plant-infecting RNA virus.[4][8]
TruD
TruD is able to modify a variety of RNA, and it is unclear how these different RNA substrates are recognized. PUS 7 modifies U2 snRNA at the position 35 and this modification will increase when the cells are in high shock. Another modification is cytoplasmic tRNA in position 13, and position 35 in pre-tRNATyr. PUS 7 modifies almost specificity does not depend on the type of RNA as mRNA show pseudouridylated by PUS 7. Recognize this the sequence of the RNA, UGUAR with the second U being the nucleotide that will be modified. The pseudouridylation of mRNA by PUS 7 increases during heat shock, because the protein moves from the nucleus to the cytoplasm. The modification is thought to increase the stability of mRNA during heat shock before the RNA goes to the nucleus or mitochondria, but more studies are needed.[4][7]
RluA
The RluA domain of these proteins can identify the substrate through a different protein binding to the substrate and then particular bonds to the RluA domain.[1][7]
PUS 5 is not well studied and located pseudouridine synthase and similar to Pus 2 does not have a mitochondrial signal targeting sequence. The protein modifies U2819 of mitochondrial 21S rRNA. Also suspected that Pus 5 modifies some Uridines in the mRNA, but again more data is needed to confirm.
PUS 6 has one that only modifies U31 of cytoplasmic and mitochondrial tRNA. Pus 6 is also known to modify mRNA.[4]
PUS 8 also known as Rib2 modifies cytoplasmic tRNA at position U32. On the C-terminus there is a DRAP-deaminase domain related to the biosynthesis of riboflavin. The RluA and DRAP or deaminase domain related to riboflavin synthase have completely separate functions in the protein and it is not known whether they interact with each other. PUS 8 is necessary in yeast, but that is suspected to be related to the riboflavin synthesis and not the pseudouridine modification.[4]
PUS 9 and PUS 8 catalyze the same position in mitochondrial tRNA instead of cytoplasmic. It is the only PUS protein that contains a mitochondrial targeting signal domain on the N-terminus. Studies suspect that PUS 9 can modify mRNAs, which would mean less substrate specificity.[4]
Techniques in genome sequencing for pseudouridine
Pseudouridine can be identified through a multitude of different techniques. A common technique to identify modifications in RNA and DNA is Liquid Chromatography with Mass Spectrometry or LC-MS. Mass spectrometry separates molecules by the mass and charge, while uridine and pseudrouidine has the same mass, but different charges. Liquid chromatography works by retention time, which has to do with leaving the column.[9] A chemical way to identify pseudouridine uses a compound called CMC or N-cyclohexyl-N′-β-(4-methylmorpholinium) ethylcarbodiimide specifically label and distinguish Uridine from Pseudouridine. CMC will bond with Pseudouridine and Uridine, but holds tighter to Pseudourdine, because of the third nitrogen able to form hydrogen bond. CMC bound to pseudouridine can then be imaged by tagging a signaling molecule. This method is still being worked to become high-throughput.[10]
Medical relevance of pseudouridine
Pseudouridine exerts a subtle but significant influence on the nearby sugar-phosphate backbone and also enhances base stacking. These effects may underlie the biological role of most, but perhaps not all of the pseudouridine residues in RNA. Certain genetic mutants lacking specific pseudouridine residues in tRNA or rRNA exhibit difficulties in translation, display slow growth rates, and fail to compete effectively with wild-type strains in mixed culture. Pseudouridine modifications are also implicated in human diseases such as mitochondrial myopathy and sideroblastic anemia (MLASA) and Dyskeratosis congenita.[4] Dyskeratosis congentia and Hoyeraal-Hreidarsson are two rare inherited syndromes caused by mutations in DKC1, the gene encoding for the pseudouridine synthase dyskerin. Pseudouridines have been recognized as regulators of viral latency processes in human immunodeficiency virus (HIV) infections.[11] Pseudouridylation has also been associated with the pathogenesis of maternally inherited diabetes and deafness (MIDD). In particular, a point mutation in a mitochondrial tRNA seems to prevent the pseudouridylation of one nucleotide, thus altering the tRNA tertiary structure. This may lead to higher tRNA instability, causing deficiencies in mitochondrial translation and respiration.[11]
See also
- Pseudouridine kinase
- TRNA-pseudouridine synthase
- PUS1
References
- Hamma, Tomoko; Ferré-D'Amaré, Adrian R. (November 2006). "Pseudouridine Synthases". Chemistry & Biology. 13 (11): 1125–1135. doi:10.1016/j.chembiol.2006.09.009. ISSN 1074-5521. PMID 17113994.
- Gray, Michael Charette, Michael W. (2000-05-01). "Pseudouridine in RNA: What, Where, How, and Why". IUBMB Life. 49 (5): 341–351. doi:10.1080/152165400410182. ISSN 1521-6543. PMID 10902565.
- Ge, Junhui; Yu, Yi-Tao (April 2013). "RNA pseudouridylation: new insights into an old modification". Trends in Biochemical Sciences. 38 (4): 210–218. doi:10.1016/j.tibs.2013.01.002. ISSN 0968-0004. PMC 3608706. PMID 23391857.
- Rintala-Dempsey, Anne C.; Kothe, Ute (2017-01-03). "Eukaryotic stand-alone pseudouridine synthases – RNA modifying enzymes and emerging regulators of gene expression?". RNA Biology. 14 (9): 1185–1196. doi:10.1080/15476286.2016.1276150. ISSN 1547-6286. PMC 5699540. PMID 28045575.
- Wu, Guowei; Radwan, Mohamed K.; Xiao, Mu; Adachi, Hironori; Fan, Jason; Yu, Yi-Tao (2016-06-07). "TheTORsignaling pathway regulates starvation-induced pseudouridylation of yeast U2 snRNA". RNA. 22 (8): 1146–1152. doi:10.1261/rna.056796.116. ISSN 1355-8382. PMC 4931107. PMID 27268497.
- Adachi, Hironori; De Zoysa, Meemanage D.; Yu, Yi-Tao (March 2019). "Post-transcriptional pseudouridylation in mRNA as well as in some major types of noncoding RNAs". Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 1862 (3): 230–239. doi:10.1016/j.bbagrm.2018.11.002. ISSN 1874-9399. PMC 6401265. PMID 30414851.
- Penzo, M.; Guerrieri, A. N.; Zacchini, F.; Treré, D.; Montanaro, L. (2017-11-01). "RNA Pseudouridylation in Physiology and Medicine: For Better and for Worse". Genes. 8 (11): 301. doi:10.3390/genes8110301. ISSN 2073-4425. PMC 5704214. PMID 29104216.
- Keffer-Wilkes, Laura Carole; Veerareddygari, Govardhan Reddy; Kothe, Ute (2016-11-14). "RNA modification enzyme TruB is a tRNA chaperone". Proceedings of the National Academy of Sciences. 113 (50): 14306–14311. doi:10.1073/pnas.1607512113. ISSN 0027-8424. PMC 5167154. PMID 27849601.
- Xu, J.; Gu, A. Y.; Thumati, N. R.; Wong JMY (2017-09-05). "Quantification of Pseudouridine Levels in Cellular RNA Pools with a Modified HPLC-UV Assay". Genes. 8 (9): 219. doi:10.3390/genes8090219. ISSN 2073-4425. PMC 5615352. PMID 28872587.
- Kalsotra, Auinash (2016-11-02). "Faculty of 1000 evaluation for Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA". doi:10.3410/f.718875945.793524920. Cite journal requires
|journal=(help) - Zhao, Yang; Karijolich, John; Glaunsinger, Britt; Zhou, Qiang (October 2016). "Pseudouridylation of 7 SK sn RNA promotes 7 SK sn RNP formation to suppress HIV ‐1 transcription and escape from latency". EMBO Reports. 17 (10): 1441–1451. doi:10.15252/embr.201642682. ISSN 1469-221X. PMC 5048380. PMID 27558685.