Protein catabolism
In molecular biology, protein catabolism is the breakdown of proteins into amino acids and simple derivative compounds, for transport into the cell through the plasma membrane and ultimately for the polymerization into new proteins via the use of ribonucleic acids (RNA) and ribosomes. Protein catabolism, which is the breakdown of macromolecules, is essentially a digestion process.
Protein catabolism is most commonly carried out by non-specific endo- and exo-proteases. However, specific proteases are used for cleaving of proteins for regulatory and protein trafficking purposes. One example is the subclass of proteolytic enzymes called oligopeptidase.
The amino acids produced by catabolism may be directly recycled to form new proteins, converted into different amino acids, or can undergo amino acid catabolism to be converted to other compounds via the Krebs cycle.[1]
Purpose
The primary reason for protein catabolism is so organisms can convert proteins into a form of energy that the body can use. To reuse their proteins, bacteria or soil microorganisms break down their proteins through protein catabolism into their individual amino acids and are used to form bacterial proteins or oxidized for energy. To convert to energy, once the proteins are broken down, they are typically deaminated (removal of an amino group) so that they can be processed into the Krebs/Citric Acid (TCA) Cycle. By proceeding into the Citric Acid Cycle, the original proteins will be converted into usable energy for the organism.[1]
There are also other processes to convert amino acids into usable molecules to enter the TCA cycle, such as transamination (transfer of amino group), decarboxylation (removal of carboxyl group), and dehydrogenation (removal of hydrogen).[1]
The proteins are digested in the intestines to produce the amino acids. The proteins are continuously being broken down and reformed, depending on the current needs that the body has. Proteins have different half-lives:[2] some have an incredibly short half-life while others have longer ones. Those with short half-lives are primarily used in metabolic pathways or processes because they help the cell adjust continuously and quickly to the changes that occur due to these processes.[3][4]
Protein degradation
The degradation of proteins occurs within the cells, as the amino acids have to pass through certain membranes before being able to be used for different processes. This first step to protein catabolism is breaking the protein down into amino acids by cleaving their peptide bonds, also known as proteolysis. The peptide bonds are broken up by the proteasome, which is able to hydrolyze the peptide bonds by using ATP energy. This process is further helped by the use of enzymes called proteases. The proteases help cleave off the remaining peptide residues to produce individual amino acids, ready to be converted into usable molecules for either glycolysis or the TCA cycle, to produce energy for the organisms, or to be used to create new proteins.[3]
Different types of proteases help cleave the proteins in different formats. There are serine, aspartate, metalloproteases, and many other classes. All use different mechanisms to cleave the peptide bonds to begin protein degradation. For example, the serine proteases, such as trypsin, engage in a nucleophilic attack on the hydroxyl oxygen of the serine on the peptide bond's carbonyl carbon in order to cleave this bond. An acyl-enzyme intermediate is created and the mechanism continues to hydrolize the other remaining linkages.[5] On the other hand, metalloproteases, such as zinc proteases, incorporate metals to break the bonds. With zinc, its active site incorporates the zinc ion, water, and histidines (which are ligands to the zinc ion). The zinc protease also engages in a nucleophilic attack but on the carbonyl carbon, using the water's oxygen atom. The active site's base helps this process along by taking a proton from that water.[6]
In certain organisms, such as bacteria, the proteins must undergo proteolysis before the amino acids can be re-polymerized into new proteins because the original proteins cannot pass through the bacterial plasma membrane, as they are too large. After the proteins are broken down into amino acids through proteolysis, these amino acids will be able to pass through the membranes of bacteria and will once again congregate to form new proteins that the bacteria needs to function.[1]
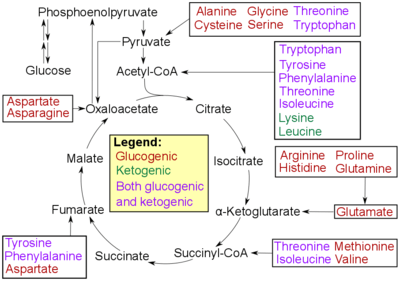
Amino Acid Degradation
Oxidative deamination is the first step to breaking down the amino acids so that they can be converted to sugars. The process begins by removing the amino group of the amino acids. The amino group becomes ammonium as it is lost and later undergoes the urea cycle to become urea, in the liver. It is then released into the blood stream, where it is transferred to the kidneys, which will secrete the urea as urine.[7][8] The remaining portion of the amino acid becomes oxidized, resulting in an alpha-keto acid. The alpha-keto acid will then proceed into the TCA cycle, in order to produce energy. The acid can also enter glycolysis, where it will be eventually converted into pyruvate. The pyruvate is then converted into acetyl-CoA so that it can enter the TCA cycle and convert the original pyruvate molecules into ATP, or usable energy for the organism.[9]
Transamination leads to the same end result as deamination: the remaining acid will undergo either glycolysis or the TCA cycle to produce energy that the organism's body will use for various purposes. This process transfers the amino group instead of losing the amino group to be converted into ammonium. The amino group is transferred to alpha-ketoglutarate, so that it can be converted to glutamate. Then glutamate transfers the amino group to oxaloacetate. This transfer is so that the oxaloacetate can be converted to aspartate or other amino acids. Eventually, this product will also proceed into oxidative deamination to once again produce alpha-ketoglutarate, an alpha-keto acid that will undergo the TCA cycle, and ammonium, which will eventually undergo the urea cycle.[3]
Transaminases are enzymes that help catalyze the reactions that take place in transamination. They help catalyze the reaction at the point when the amino group is transferred from the original amino acid, like glutamate to alpha-ketoglutarate, and hold onto it to transfer it to another alpha-ketoacid.[3]
Factors Determining Overall Rate
Some key factors that determine overall rate include protein half-life, pH, and temperature.
Protein half-life helps determine the overall rate as this designates the first step in protein catabolism. Depending on whether this step is short or long will influence the rest of the metabolic process. One key component in determining the protein half-life is based on the N-end rule. This states that the amino acid present at the N-terminus of a protein helps determine the protein's half-life.[10]
Changes in the pH and temperature of the molecular environment can also help determine the overall rate. The process that cleaves the protein's peptide bonds, proteolysis, is sensitive to changes in pH and temperature. In low pH and high temperatures, proteolysis can begin even without an enzyme. This would help speed up the overall rate since it yields the same results as adding an enzyme but without necessitating enzyme use.[11]
References
- Bauman, Robert W.; Machunis-Masuoka, Elizabeth; Tizard, Ian R. (2004-01-01). Microbiology. Pearson/Benjamin Cummings. ISBN 9780805376524.
- Zhou, Pengbo (2004-01-01). "Determining Protein Half-Lives". In Dickson, RobertC.; Mendenhall, MichaelD. (eds.). Signal Transduction Protocols. Methods in Molecular Biology. 284. Humana Press. pp. 67–77. doi:10.1385/1-59259-816-1:067. ISBN 9781588292452. PMID 15173609.
- Miles, Bryant (April 9, 2003). "Protein Catabolism" (PDF). Archived from the original (PDF) on August 12, 2014.
- Bojkowska, Karolina; Santoni de Sio, Francesca; Barde, Isabelle; Offner, Sandra; Verp, Sonia; Heinis, Christian; Johnsson, Kai; Trono, Didier (2011-06-24). "Measuring In Vivo Protein Half-Life". Chemistry & Biology. 18 (6): 805–815. doi:10.1016/j.chembiol.2011.03.014. PMID 21700215.
- Voet, D. (2004-01-01). D. Voet's Biochemistry 3rd (3 ed.). Wiley.
- Erez, Elinor; Fass, Deborah; Bibi, Eitan (2009). "How intramembrane proteases bury hydrolytic reactions in the membrane". Nature. 459 (7245): 371–378. doi:10.1038/nature08146. PMID 19458713.
- "26.9: The Catabolism of Proteins". Chemistry LibreTexts. 2014-06-19. Retrieved 2016-10-25.
- "Oxidative Deamination". chemistry.elmhurst.edu. Retrieved 2016-10-25.
- "GLYCOLYSIS AND THE KREBS CYCLE". homepage.smc.edu. Retrieved 2016-11-08.
- Tasaki, Takafumi; Sriram, Shashikanth M.; Park, Kyong Soo; Kwon, Yong Tae (2012-06-04). "The N-End Rule Pathway". Annual Review of Biochemistry. 81 (1): 261–289. doi:10.1146/annurev-biochem-051710-093308. ISSN 0066-4154. PMC 3610525. PMID 22524314.
- de Giori, G.S.; Valdez, G.F. de; Holgado, A.P. de Ruiz; Oliver, G. (1985). "Effect of pH and Temperature on the Proteolytic Activity of Lactic Acid Bacteria". Journal of Dairy Science. 68 (9): 2160–2164. doi:10.3168/jds.s0022-0302(85)81085-7.