Prophage
A prophage is a bacteriophage (often shortened to "phage") genome inserted and integrated into the circular bacterial DNA chromosome or exists as an extrachromosomal plasmid. This is a latent form of a phage, in which the viral genes are present in the bacterium without causing disruption of the bacterial cell. Pro means ''before'', so, prophage means the stage of a virus in the form of genome inserted into host DNA before being activated inside the host.
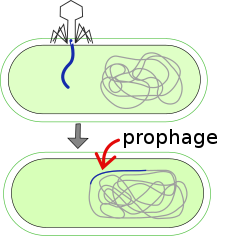
Background
Prophages are able to do a multitude of things within their respective bacterial strains. Prophages can increase the virulence potential of bacterial strains in both humans and plant pathogens as well as increase the ability of the bacteria to survive in harsh environments.[1] Pathogens have been able to adapt and thrive in a wide range of environments. Some anaerobic pathogens such as Clostridium perfringens and Clostridium difficile exist in the intestines and are unable to survive in places with large amounts of oxygen for extended periods of time.[2] Still others can reside in the soil such as B. anthracis, while pathogens such as C. difficile can even survive in very sterile hospital settings. Prophages can provide these bacteria with both resistance mechanisms as well as metabolic advantages which give the host cell the best chance of survival,[3] sometimes even completely altering the bacterial genome.[4]
Prophage induction
Upon detection of host cell damage by UV light or certain chemicals, the prophage is excised from the bacterial chromosome in a process called prophage induction. After induction, viral replication begins via the lytic cycle. In the lytic cycle, the virus commandeers the cell's reproductive machinery. The cell may fill with new viruses until it lyses or bursts, or it may release the new viruses one at a time in an exocytotic process. The period from infection to lysis is termed the latent period. A virus following a lytic cycle is called a virulent virus. Prophages are important agents of horizontal gene transfer, and are considered part of the mobilome. All families of bacterial viruses that have circular (single-stranded or double-stranded) DNA genomes or replicate their genomes through rolling circle replication (e.g., Caudovirales) have temperate members.[5]
Zygotic induction
Zygotic induction occurs when a bacterial cell carrying the DNA of a bacterial virus transfers its own DNA along with the viral DNA (prophage) into the new host cell. This has the effect of causing the host cell to break apart.[6] The DNA of the bacterial cell is silenced before entry into the cell by a repressor protein which is encoded for by the prophage. Upon the transfer of the bacterial cell's DNA into the host cell, the repressor protein is no longer encoded for, and the bacterial cell's original DNA is then turned on in the host cell. This mechanism eventually will lead to the release of the virus as the host cell splits open and the viral DNA is able to spread.[7] This new discovery provided key insights into bacterial conjugation and contributed to the early repression model of gene regulation, which provided an explanation as to how the lac operon and λ bacteriophage genes are negatively regulated.
Prophage reactivation
Bacteriophage λ is able to undergo a type of recombinational repair called prophage reactivation.[8][9] Prophage reactivation can occur by recombination between a UV-damaged infecting phage λ chromosome and a homologous phage genome integrated into the bacterial DNA and existing in a prophage state. Prophage reactivation in the case of phage λ appears to be an accurate recombinational repair process that is mediated by the recA+ and red+ gene products.
Applications
Prophages can tell researchers a lot about the relationship between a bacterium and a host.[10] With data from more nonpathogenic bacteria, researchers will be able to gather evidence as to whether or not prophages contribute to the survival value of the host. Prophage genomics has the potential to lead to ecological adaptations of the relationships between bacteria.[11] Another important area of interest is the control of prophage gene expression with many of the lysogenic conversion genes (gene conversion) being tightly regulated.[12] This process is capable of converting non-pathogenic bacteria into pathogenic bacteria that can now produce harmful toxins[13] such as in staph infections. Since the specific mechanisms of prophage are not yet detailed, this research could provide the community with this tool for future research.[14]
References
- Menouni R, Hutinet G, Petit MA, Ansaldi M (2015). "Bacterial genome remodeling through bacteriophage recombination". FEMS Microbiology Letters. 362 (1): 1–10. doi:10.1093/femsle/fnu022.CS1 maint: uses authors parameter (link)
- Fortier L.C, Sekulovic O (2013). "Importance of prophages to evolution and virulence of bacterial pathogens". Virulence. 4 (5): 354–365. doi:10.4161/viru.24498.CS1 maint: uses authors parameter (link)
- Fortier L.C, Sekulovic O (2013). "Importance of prophages to evolution and virulence of bacterial pathogens". Virulence. 4 (5): 354–365. doi:10.4161/viru.24498.CS1 maint: uses authors parameter (link)
- Menouni R, Hutinet G, Petit MA, Ansaldi M (2015). "Bacterial genome remodeling through bacteriophage recombination". FEMS Microbiology Letters. 362 (1): 1–10. doi:10.1093/femsle/fnu022.CS1 maint: uses authors parameter (link)
- Krupovic M, Prangishvili D, Hendrix RW, Bamford DH (2011). "Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere". Microbiol Mol Biol Rev. 75 (4): 610–635. doi:10.1128/MMBR.00011-11. PMC 3232739. PMID 22126996.CS1 maint: uses authors parameter (link)
- Griffiths A, Miller J, Suzuki D, Lewontin R, Gelbart W (2016). "An Introduction to Genetic Analysis". NCBI bookshelf.CS1 maint: uses authors parameter (link)
- Griffiths A, Miller J, Suzuki D, Lewontin R, Gelbart W (2016). "An Introduction to Genetic Analysis". NCBI bookshelf.CS1 maint: uses authors parameter (link)
- Blanco M, Devoret R (March 1973). "Repair mechanisms involved in prophage reactivation and UV reactivation of UV-irradiated phage lambda". Mutation Research. 17 (3): 293–305. doi:10.1016/0027-5107(73)90001-8. PMID 4688367
- Bernstein C. Deoxyribonucleic acid repair in bacteriophage. Microbiol Rev. 1981;45(1):72-98
- Canchaya C, Proux C, Fournous G, Bruttin A, Brüssow H (2003). "Prophage Genomics". Microbiology and Molecular Biology Reviews. 67 (2): 238–276. doi:10.1128/MMBR.67.2.238-276.2003.CS1 maint: uses authors parameter (link)
- Canchaya C, Proux C, Fournous G, Bruttin A, Brüssow H (2003). "Prophage Genomics". Microbiology and Molecular Biology Reviews. 67 (2): 238–276. doi:10.1128/MMBR.67.2.238-276.2003.CS1 maint: uses authors parameter (link)
- Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits A (2015). "A new perspective on lysogeny: prophages as active regulatory switches of bacteria". Nature Reviews Microbiology. 13 (10): 641–650. doi:10.1038/nrmicro3527. PMID 26373372.CS1 maint: uses authors parameter (link)
- Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits A (2015). "A new perspective on lysogeny: prophages as active regulatory switches of bacteria". Nature Reviews Microbiology. 13 (10): 641–650. doi:10.1038/nrmicro3527. PMID 26373372.CS1 maint: uses authors parameter (link)
- Canchaya C, Proux C, Fournous G, Bruttin A, Brüssow H (2003). "Prophage Genomics". Microbiology and Molecular Biology Reviews. 67 (2): 238–276. doi:10.1128/MMBR.67.2.238-276.2003.CS1 maint: uses authors parameter (link)