PIN1
Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 is an enzyme that in humans is encoded by the PIN1 gene.[4][5]
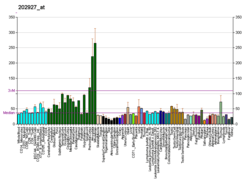
Pin 1, or peptidyl-prolyl cis/trans isomerase (PPIase), isomerizes only phospho-Serine/Threonine-Proline motifs. The enzyme binds to a subset of proteins and thus plays a role as a post phosphorylation control in regulating protein function. Studies have shown that the deregulation of Pin1 may play a pivotal role in various diseases. Notably, the up-regulation of Pin1 is implicated in certain cancers, and the down-regulation of Pin1 is implicated in Alzheimer's disease. Inhibitors of Pin1 may have therapeutic implications for cancer[6][7] and immune disorders.[8]
Discovery
The gene encoding Pin1 was identified in 1996 as a result of a genetic/biochemical screen for proteins involved in mitotic regulation. It was found to be essential for cell division in some organisms. By 1999, however, it was apparent that Pin1 knockout mice had a surprisingly mild phenotype, indicating that the enzyme was not required for cell division per se. Further studies later found that loss of Pin1 in mice displays are not only neuronal degenerative phenotypes but also several abnormalities, similar to those of cyclin D1-null mice, suggesting the conformation changes mediated by Pin1 may be crucial for cell normal function.
Activation
Phosphorylation of Ser/Thr-Pro motifs in substrates is required for recognition by Pin1. Pin is a small protein at 18 kDa and does not have a nuclear localization or export signal. However, 2009, Lufei et al. reported that Pin1 has putative novel nuclear localization signal (NLS) and Pin1 interacts with importin α5 (KPNA1).[9] Substrate interactions and a WW domain determine subcellular distribution. Expression is induced by growth signals from E2F transcription factors. Expression levels fluctuate in normal, but not in cancerous cells. Expression is often associated with cell proliferation. Postranslational modifications such as phosphorylation on Ser16 inhibit the ability of Pin1 to bind substrate, and this inhibitory process may be altered during oncogenesis. It is hypothesized, but not proven, that Pin1 might also be regulated by proteolytic pathways.
Function
Pin1 activity regulates the outcome of proline-directed kinase (e.g. MAPK, CDK or GSK3) signalling and consequently regulates cell proliferation (in part through control of cyclin D1 levels and stability) and cell survival. The precise effects of Pin1 depend upon the system: Pin1 accelerates dephosphorylation of Cdc25 and Tau, but protects phosphorylated cyclin D from ubiquitination and proteolysis. Recent data also implicate Pin1 as playing an important role in immune responses, at least in part by increasing the stability of cytokine mRNAs by influencing the protein complexes to which they bind. Pin1 has been hypothesized to act as a molecular timer.[10]
Interactions
PIN1 has been shown to interact with:
- C-jun,[11]
- CDC25C,[12][13][14]
- CDC27,[12][14]
- CSNK2A2,[15]
- Casein kinase 2, alpha 1,[15]
- DAB2,[16]
- eNOS,[17]
- FOXO4,[18]
- MPHOSPH1,[19]
- MYT1,[20]
- Mothers against decapentaplegic homolog 2,[21]
- Mothers against decapentaplegic homolog 3[21]
- P53,[22][23]
- PKMYT1,[12]
- PLK1,[12][14]
- SUPT5H,[24] and
- Wee1-like protein kinase.[12]
References
- GRCh38: Ensembl release 89: ENSG00000127445 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Lu KP, Hanes SD, Hunter T (Apr 1996). "A human peptidyl-prolyl isomerase essential for regulation of mitosis". Nature. 380 (6574): 544–7. doi:10.1038/380544a0. PMID 8606777.
- "Entrez Gene: PIN1 Protein (peptidylprolyl cis/trans isomerase) NIMA-interacting 1".
- da Costa, Kauê Santana; Galúcio, João Marcos; de Jesus, Deivid Almeida; Gomes, Guelber Cardoso; Lima e Lima, Anderson Henrique; Taube, Paulo Sérgio; dos Santos, Alberto Monteiro; Lameira, Jerônimo (2019-10-25). "Targeting Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1: A Structure-based Virtual Screening Approach to Find Novel Inhibitors". Current Computer-Aided Drug Design. 15. doi:10.2174/1573409915666191025114009. PMID 31654518.
- Campaner, Elena; Rustighi, Alessandra; Zannini, Alessandro; Cristiani, Alberto; Piazza, Silvano; Ciani, Yari; Kalid, Ori; Golan, Gali; Baloglu, Erkan; Shacham, Sharon; Valsasina, Barbara (August 2017). "A covalent PIN1 inhibitor selectively targets cancer cells by a dual mechanism of action". Nature Communications. 8 (1): 15772. doi:10.1038/ncomms15772. ISSN 2041-1723. PMC 5472749. PMID 28598431.
- Rudrabhatla, P.; Albers, W.; Pant, H. C. (2009-11-25). "Peptidyl-Prolyl Isomerase 1 Regulates Protein Phosphatase 2A-Mediated Topographic Phosphorylation of Neurofilament Proteins". Journal of Neuroscience. 29 (47): 14869–14880. doi:10.1523/JNEUROSCI.4469-09.2009. ISSN 0270-6474. PMC 3849796. PMID 19940183.
- Lufei C, Cao X (2009). "Nuclear import of Pin1 is mediated by a novel sequence in the PPIase domain". FEBS Letters. 583 (2): 271–276. doi:10.1016/j.febslet.2008.12.011. PMID 19084525.
- Lu KP, Finn G, Lee TH, Nicholson LK (Oct 2007). "Prolyl cis-trans isomerization as a molecular timer". Nature Chemical Biology. 3 (10): 619–29. doi:10.1038/nchembio.2007.35. PMID 17876319.
- Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, Lu KP (Jul 2001). "Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1". The EMBO Journal. 20 (13): 3459–72. doi:10.1093/emboj/20.13.3459. PMC 125530. PMID 11432833.
- Shen M, Stukenberg PT, Kirschner MW, Lu KP (Mar 1998). "The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins". Genes & Development. 12 (5): 706–20. doi:10.1101/gad.12.5.706. PMC 316589. PMID 9499405.
- Goldstrohm AC, Albrecht TR, Suñé C, Bedford MT, Garcia-Blanco MA (Nov 2001). "The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1". Molecular and Cellular Biology. 21 (22): 7617–28. doi:10.1128/MCB.21.22.7617-7628.2001. PMC 99933. PMID 11604498.
- Lu PJ, Zhou XZ, Shen M, Lu KP (Feb 1999). "Function of WW domains as phosphoserine- or phosphothreonine-binding modules". Science. 283 (5406): 1325–8. doi:10.1126/science.283.5406.1325. PMID 10037602.
- Messenger MM, Saulnier RB, Gilchrist AD, Diamond P, Gorbsky GJ, Litchfield DW (Jun 2002). "Interactions between protein kinase CK2 and Pin1. Evidence for phosphorylation-dependent interactions". The Journal of Biological Chemistry. 277 (25): 23054–64. doi:10.1074/jbc.M200111200. PMID 11940573.
- He J, Xu J, Xu XX, Hall RA (Jul 2003). "Cell cycle-dependent phosphorylation of Disabled-2 by cdc2". Oncogene. 22 (29): 4524–30. doi:10.1038/sj.onc.1206767. PMID 12881709.
- Ruan L, Torres CM, Qian J, Chen F, Mintz JD, Stepp DW, Fulton D, Venema RC (Feb 2011). "Pin1 prolyl isomerase regulates endothelial nitric oxide synthase". Arteriosclerosis, Thrombosis, and Vascular Biology. 31 (2): 392–8. doi:10.1161/ATVBAHA.110.213181. PMC 3075952. PMID 21051667.
- Brenkman AB, de Keizer PL, van den Broek NJ, van der Groep P, van Diest PJ, van der Horst A, Smits AM, Burgering BM (Sep 2008). "The peptidyl-isomerase Pin1 regulates p27kip1 expression through inhibition of Forkhead box O tumor suppressors". Cancer Research. 68 (18): 7597–605. doi:10.1158/0008-5472.CAN-08-1059. PMID 18794148.
- Kamimoto T, Zama T, Aoki R, Muro Y, Hagiwara M (Oct 2001). "Identification of a novel kinesin-related protein, KRMP1, as a target for mitotic peptidyl-prolyl isomerase Pin1". The Journal of Biological Chemistry. 276 (40): 37520–8. doi:10.1074/jbc.M106207200. PMID 11470801.
- Wells NJ, Watanabe N, Tokusumi T, Jiang W, Verdecia MA, Hunter T (Oct 1999). "The C-terminal domain of the Cdc2 inhibitory kinase Myt1 interacts with Cdc2 complexes and is required for inhibition of G(2)/M progression". Journal of Cell Science. 112 (19): 3361–71. PMID 10504341.
- Nakano A, Koinuma D, Miyazawa K, Uchida T, Saitoh M, Kawabata M, Hanai J, Akiyama H, Abe M, Miyazono K, Matsumoto T, Imamura T (Mar 2009). "Pin1 down-regulates transforming growth factor-beta (TGF-beta) signaling by inducing degradation of Smad proteins". The Journal of Biological Chemistry. 284 (10): 6109–15. doi:10.1074/jbc.M804659200. PMID 19122240.
- Wulf GM, Liou YC, Ryo A, Lee SW, Lu KP (Dec 2002). "Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage". The Journal of Biological Chemistry. 277 (50): 47976–9. doi:10.1074/jbc.C200538200. PMID 12388558.
- Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S, Ronai Z, Blandino G, Schneider C, Del Sal G (Oct 2002). "The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults". Nature. 419 (6909): 853–7. doi:10.1038/nature01120. PMID 12397362.
- Lavoie SB, Albert AL, Handa H, Vincent M, Bensaude O (Sep 2001). "The peptidyl-prolyl isomerase Pin1 interacts with hSpt5 phosphorylated by Cdk9". Journal of Molecular Biology. 312 (4): 675–85. doi:10.1006/jmbi.2001.4991. PMID 11575923.
Further reading
- Lu KP, Liou YC, Zhou XZ (Apr 2002). "Pinning down proline-directed phosphorylation signaling". Trends in Cell Biology. 12 (4): 164–72. doi:10.1016/S0962-8924(02)02253-5. PMID 11978535.
- Wulf G, Finn G, Suizu F, Lu KP (May 2005). "Phosphorylation-specific prolyl isomerization: is there an underlying theme?". Nature Cell Biology. 7 (5): 435–41. doi:10.1038/ncb0505-435. PMID 15867923.
- Etzkorn FA (May 2006). "Pin1 flips Alzheimer's switch". ACS Chemical Biology. 1 (4): 214–6. doi:10.1021/cb600171g. PMID 17163675.
- Balastik M, Lim J, Pastorino L, Lu KP (Apr 2007). "Pin1 in Alzheimer's disease: multiple substrates, one regulatory mechanism?". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1772 (4): 422–9. doi:10.1016/j.bbadis.2007.01.006. PMC 1868500. PMID 17317113.
- Maleszka R, Hanes SD, Hackett RL, de Couet HG, Miklos GL (Jan 1996). "The Drosophila melanogaster dodo (dod) gene, conserved in humans, is functionally interchangeable with the ESS1 cell division gene of Saccharomyces cerevisiae". Proceedings of the National Academy of Sciences of the United States of America. 93 (1): 447–51. doi:10.1073/pnas.93.1.447. PMC 40255. PMID 8552658.
- Ranganathan R, Lu KP, Hunter T, Noel JP (Jun 1997). "Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent". Cell. 89 (6): 875–86. doi:10.1016/S0092-8674(00)80273-1. PMID 9200606.
- Campbell HD, Webb GC, Fountain S, Young IG (Sep 1997). "The human PIN1 peptidyl-prolyl cis/trans isomerase gene maps to human chromosome 19p13 and the closely related PIN1L gene to 1p31". Genomics. 44 (2): 157–62. doi:10.1006/geno.1997.4854. PMID 9299231.
- Crenshaw DG, Yang J, Means AR, Kornbluth S (Aug 1998). "The mitotic peptidyl-prolyl isomerase, Pin1, interacts with Cdc25 and Plx1". The EMBO Journal. 17 (5): 1315–27. doi:10.1093/emboj/17.5.1315. PMC 1170480. PMID 9482729.
- Shen M, Stukenberg PT, Kirschner MW, Lu KP (Mar 1998). "The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins". Genes & Development. 12 (5): 706–20. doi:10.1101/gad.12.5.706. PMC 316589. PMID 9499405.
- Lu PJ, Zhou XZ, Shen M, Lu KP (Feb 1999). "Function of WW domains as phosphoserine- or phosphothreonine-binding modules". Science. 283 (5406): 1325–8. doi:10.1126/science.283.5406.1325. PMID 10037602.
- Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP (Jun 1999). "The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein". Nature. 399 (6738): 784–8. doi:10.1038/21650. PMID 10391244.
- Albert A, Lavoie S, Vincent M (Aug 1999). "A hyperphosphorylated form of RNA polymerase II is the major interphase antigen of the phosphoprotein antibody MPM-2 and interacts with the peptidyl-prolyl isomerase Pin1". Journal of Cell Science. 112. 112 (15): 2493–500. PMID 10393805.
- Wells NJ, Watanabe N, Tokusumi T, Jiang W, Verdecia MA, Hunter T (Oct 1999). "The C-terminal domain of the Cdc2 inhibitory kinase Myt1 interacts with Cdc2 complexes and is required for inhibition of G(2)/M progression". Journal of Cell Science. 112. 112 (19): 3361–71. PMID 10504341.
- Gerez L, Mohrmann K, van Raak M, Jongeneelen M, Zhou XZ, Lu KP, van Der Sluijs P (Jul 2000). "Accumulation of rab4GTP in the cytoplasm and association with the peptidyl-prolyl isomerase pin1 during mitosis". Molecular Biology of the Cell. 11 (7): 2201–11. doi:10.1091/mbc.11.7.2201. PMC 14913. PMID 10888662.
- Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP (Aug 2000). "Structural basis for phosphoserine-proline recognition by group IV WW domains". Nature Structural Biology. 7 (8): 639–43. doi:10.1038/77929. PMID 10932246.
- Rippmann JF, Hobbie S, Daiber C, Guilliard B, Bauer M, Birk J, Nar H, Garin-Chesa P, Rettig WJ, Schnapp A (Jul 2000). "Phosphorylation-dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis". Cell Growth & Differentiation. 11 (7): 409–16. PMID 10939594.
- Liu W, Youn HD, Zhou XZ, Lu KP, Liu JO (May 2001). "Binding and regulation of the transcription factor NFAT by the peptidyl prolyl cis-trans isomerase Pin1". FEBS Letters. 496 (2–3): 105–8. doi:10.1016/S0014-5793(01)02411-5. PMID 11356192.
- Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, Lu KP (Jul 2001). "Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1". The EMBO Journal. 20 (13): 3459–72. doi:10.1093/emboj/20.13.3459. PMC 125530. PMID 11432833.
- Kamimoto T, Zama T, Aoki R, Muro Y, Hagiwara M (Oct 2001). "Identification of a novel kinesin-related protein, KRMP1, as a target for mitotic peptidyl-prolyl isomerase Pin1". The Journal of Biological Chemistry. 276 (40): 37520–8. doi:10.1074/jbc.M106207200. PMID 11470801.