Phagocytosis
Phagocytosis (from Ancient Greek φαγεῖν (phagein) , meaning 'to eat', and κύτος, (kytos) , meaning 'cell') is the process by which a cell uses its plasma membrane to engulf a large particle (≥ 0.5 μm), giving rise to an internal compartment called the phagosome. It is one type of endocytosis.
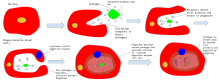
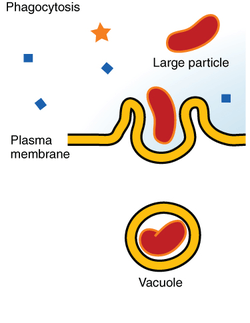
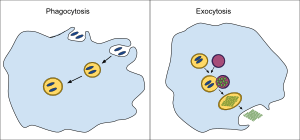
In a multicellular organism's immune system, phagocytosis is a major mechanism used to remove pathogens and cell debris. The ingested material is then digested in the phagosome. Bacteria, dead tissue cells, and small mineral particles are all examples of objects that may be phagocytized. Some protozoa use phagocytosis as means to obtain nutrients.
History
Phagocytosis was first noted by Canadian physician William Osler (1876),[1] and later studied and named by Élie Metchnikoff (1880, 1883).[2]
In immune system
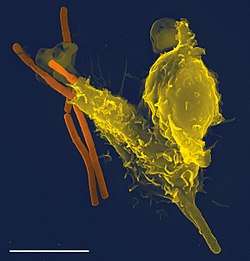
Phagocytosis is one of the main mechanisms of the innate immune defense. It is one of the first processes responding to infection, and is also one of the initiating branches of an adaptive immune response. Although most cells are capable of phagocytosis, some cell types perform it as part of their main function. These are called 'professional phagocytes.' Phagocytosis is old in evolutionary terms, being present even in invertebrates.[3]
Professional phagocytic cells
Neutrophils, macrophages, monocytes, dendritic cells, osteoclasts and eosinophils can be classified as professional phagocytes.[2] The first three have the greatest role in immune response to most infections.[3]
The role of neutrophils is patrolling the bloodstream and rapid migration to the tissues in large numbers only in case of infection.[3] There they have direct microbicidal effect by phagocytosis. After ingestion, neutrophils are efficient in intracellular killing of pathogens. Neutrophils phagocytose mainly via the Fcγ receptors and complement receptors 1 and 3. The microbicidal effect of neutrophils is due to a large repertoire of molecules present in pre-formed granules. Enzymes and other molecules prepared in these granules are proteases, such as collagenase, gelatinase or serine proteases, myeloperoxidase, lactoferrin and antibiotic proteins. Degranulation of these into the phagosome, accompanied by high reactive oxygen species production (oxidative burst) is highly microbicidal.[4]
Monocytes, and the macrophages that mature from them, leave blood circulation to migrate through tissues. There they are resident cells and form a resting barrier.[3] Macrophages initiate phagocytosis by mannose receptors, scavenger receptors, Fcγ receptors and complement receptors 1, 3 and 4. Macrophages are long-lived and can continue phagocytosis by forming new lysosomes.[3][5]
Dendritic cells also reside in tissues and ingest pathogens by phagocytosis. Their role is not killing or clearance of microbes, but rather breaking them down for antigen presentation to the cells of the adaptive immune system.[3]
Initiating receptors
Receptors for phagocytosis can be divided into two categories by recognised molecules. The first, opsonic receptors, are dependent on opsonins.[6] Among these are receptors that recognise the Fc part of bound IgG antibodies, deposited complement or receptors, that recognise other opsonins of cell or plasma origin. Non-opsonic receptors include lectin-type receptors, Dectin receptor, or scavenger receptors. Some phagocytic pathways require a second signal from pattern recognition receptors (PRRs) activated by attachment to pathogen-associated molecular patterns (PAMPS), which leads to NF-κB activation.[2]
Fcγ receptors
Fcγ receptors recognise IgG coated targets. The main recognised part is the Fc fragment. The molecule of the receptor contain an intracellular ITAM domain or associates with an ITAM-containing adaptor molecule. ITAM domains transduce the signal from the surface of the phagocyte to the nucleus. For example, activating receptors of human macrophages are FcγRI, FcγRIIA, and FcγRIII.[5] Fcγ receptor mediated phagocytosis includes formation of protrusions of the cell called a 'phagocytic cup' and activates an oxidative burst in neutrophils.[4]
Complement receptors
These receptors recognise targets coated in C3b, C4b and C3bi from plasma complement. The extracellular domain of the receptors contains a lectin-like complement-binding domain. Recognition by complement receptors is not enough to cause internalisation without additional signals. In macrophages, the CR1, CR3 and CR4 are responsible for recognition of targets. Complement coated targets are internalised by 'sinking' into the phagocyte membrane, without any protrusions.[5]
Mannose receptors
Mannose and other pathogen-associated sugars, such as fucose, are recognised by the mannose receptor. Eight lectin-like domains form the extracellular part of the receptor. The ingestion mediated by the mannose receptor is distinct in molecular mechanisms from Fcγ receptor or complement receptor mediated phagocytosis.[5]
Phagosome
Engulfment of material is facilitated by the actin-myosin contractile system. The phagosome is the organelle formed by phagocytosis of material. It then moves toward the centrosome of the phagocyte and is fused with lysosomes, forming a phagolysosome and leading to degradation. Progressively, the phagolysosome is acidified, activating degradative enzymes.[2][7]
Degradation can be oxygen-dependent or oxygen-independent.
- Oxygen-dependent degradation depends on NADPH and the production of reactive oxygen species. Hydrogen peroxide and myeloperoxidase activate a halogenating system, which leads to the creation of hypochlorite and the destruction of bacteria.[8]
- Oxygen-independent degradation depends on the release of granules, containing enzymes such as lysozymes, and cationic proteins such as defensins. Other antimicrobial peptides are present in these granules, including lactoferrin, which sequesters iron to provide unfavourable growth conditions for bacteria. Other enzymes like hyaluronidase, lipase, collagenase, elastase, ribonuclease, deoxyribonuclease also play an important role in preventing the spread of infection and degradation of essential microbial biomolecules leading to cell death.[4][5]
Leukocytes generate hydrogen cyanide during phagocytosis, and can kill bacteria, fungi, and other pathogens by generating several other toxic chemicals.[9][10][11]
Some bacteria, for example Treponema pallidum, Escheria coli and Staphylococcus aureus, are able to avoid phagocytosis by several mechanisms.
In apoptosis
Following apoptosis, the dying cells need to be taken up into the surrounding tissues by macrophages in a process called efferocytosis. One of the features of an apoptotic cell is the presentation of a variety of intracellular molecules on the cell surface, such as calreticulin, phosphatidylserine (from the inner layer of the plasma membrane), annexin A1, oxidised LDL and altered glycans.[12] These molecules are recognised by receptors on the cell surface of the macrophage such as the phosphatidylserine receptor or by soluble (free-floating) receptors such as thrombospondin 1, GAS6, and MFGE8, which themselves then bind to other receptors on the macrophage such as CD36 and alpha-v beta-3 integrin. Defects in apoptotic cell clearance is usually associated with impaired phagocytosis of macrophages. Accumulation of apoptotic cell remnants often causes autoimmune disorders; thus pharmacological potentiation of phagocytosis has a medical potential in treatment of certain forms of autoimmune disorders.[13][14][15][16]
In protists
In many protists, phagocytosis is used as a means of feeding, providing part or all of their nourishment. This is called phagotrophic nutrition, distinguished from osmotrophic nutrition which takes place by absorption.
- In some, such as amoeba, phagocytosis takes place by surrounding the target object with pseudopods, as in animal phagocytes. In humans, the amoebozoan Entamoeba histolytica can phagocytose red blood cells.
- Ciliates also engage in phagocytosis.[17] In ciliates there is a specialized groove or chamber in the cell where phagocytosis takes place, called the cytostome or mouth.
As in phagocytic immune cells, the resulting phagosome may be merged with lysosomes containing digestive enzymes, forming a phagolysosome. The food particles will then be digested, and the released nutrients are diffused or transported into the cytosol for use in other metabolic processes.[18]
Mixotrophy can involve phagotrophic nutrition and phototrophic nutrition.[19]
See also
References
- Ambrose, Charles T. (2006). "The Osler slide, a demonstration of phagocytosis from 1876: Reports of phagocytosis before Metchnikoff's 1880 paper". Cellular Immunology. 240 (1): 1–4. doi:10.1016/j.cellimm.2006.05.008. PMID 16876776.
- Gordon, Siamon (March 2016). "Phagocytosis: An Immunobiologic Process". Immunity. 44 (3): 463–475. doi:10.1016/j.immuni.2016.02.026. PMID 26982354.
- M.), Murphy, Kenneth (Kenneth (2012). Janeway's immunobiology. Travers, Paul, 1956-, Walport, Mark., Janeway, Charles. (8th ed.). New York: Garland Science. ISBN 9780815342434. OCLC 733935898.
- Witko-Sarsat, Véronique; Rieu, Philippe; Descamps-Latscha, Béatrice; Lesavre, Philippe; Halbwachs-Mecarelli, Lise (May 2000). "Neutrophils: Molecules, Functions and Pathophysiological Aspects". Laboratory Investigation. 80 (5): 617–653. doi:10.1038/labinvest.3780067. ISSN 0023-6837. PMID 10830774.
- Aderem, Alan; Underhill, David M. (April 1999). "Mechanisms of Phagocytosis in Macrophages". Annual Review of Immunology. 17 (1): 593–623. doi:10.1146/annurev.immunol.17.1.593. ISSN 0732-0582. PMID 10358769.
- The Immune System, Peter Parham, Garland Science, 2nd edition
- Flannagan, Ronald S.; Jaumouillé, Valentin; Grinstein, Sergio (2012-02-28). "The Cell Biology of Phagocytosis". Annual Review of Pathology: Mechanisms of Disease. 7 (1): 61–98. doi:10.1146/annurev-pathol-011811-132445. ISSN 1553-4006. PMID 21910624.
- "Archived copy" (PDF). Archived from the original (PDF) on 2016-03-03. Retrieved 2011-10-28.CS1 maint: archived copy as title (link)
- Borowitz JL, Gunasekar PG, Isom GE (12 Sep 1997). "Hydrogen cyanide generation by mu-opiate receptor activation: possible neuromodulatory role of endogenous cyanide". Brain Research. 768 (1–2): 294–300. doi:10.1016/S0006-8993(97)00659-8. PMID 9369328.
- Stelmaszyńska, T (1985). "Formation of HCN by human phagocytosing neutrophils--1. Chlorination of Staphylococcus epidermidis as a source of HCN". Int J Biochem. 17 (3): 373–9. doi:10.1016/0020-711x(85)90213-7. PMID 2989021.
- Zgliczyński, Jan Maciej; Stelmaszyńska, Teresa (1988). The Respiratory Burst and its Physiological Significance. pp. 315–347. doi:10.1007/978-1-4684-5496-3_15. ISBN 978-1-4684-5498-7.
- Bilyy RO, Shkandina T, Tomin A, Muñoz LE, Franz S, Antonyuk V, Kit YY, Zirngibl M, Fürnrohr BG, Janko C, Lauber K, Schiller M, Schett G, Stoika RS, Herrmann M (January 2012). "Macrophages discriminate glycosylation patterns of apoptotic cell-derived microparticles". The Journal of Biological Chemistry. 287 (1): 496–503. doi:10.1074/jbc.M111.273144. PMC 3249103. PMID 22074924.
- Mukundan L, Odegaard JI, Morel CR, Heredia JE, Mwangi JW, Ricardo-Gonzalez RR, Goh YP, Eagle AR, Dunn SE, Awakuni JU, Nguyen KD, Steinman L, Michie SA, Chawla A (November 2009). "PPAR-delta senses and orchestrates clearance of apoptotic cells to promote tolerance". Nature Medicine. 15 (11): 1266–72. doi:10.1038/nm.2048. PMC 2783696. PMID 19838202.
- Roszer, T; Menéndez-Gutiérrez, MP; Lefterova, MI; Alameda, D; Núñez, V; Lazar, MA; Fischer, T; Ricote, M (Jan 1, 2011). "Autoimmune kidney disease and impaired engulfment of apoptotic cells in mice with macrophage peroxisome proliferator-activated receptor gamma or retinoid X receptor alpha deficiency". Journal of Immunology. 186 (1): 621–31. doi:10.4049/jimmunol.1002230. PMC 4038038. PMID 21135166.
- Kruse, K; Janko, C; Urbonaviciute, V; Mierke, CT; Winkler, TH; Voll, RE; Schett, G; Muñoz, LE; Herrmann, M (September 2010). "Inefficient clearance of dying cells in patients with SLE: anti-dsDNA autoantibodies, MFG-E8, HMGB-1 and other players". Apoptosis. 15 (9): 1098–113. doi:10.1007/s10495-010-0478-8. PMID 20198437.
- Han, CZ; Ravichandran, KS (Dec 23, 2011). "Metabolic connections during apoptotic cell engulfment". Cell. 147 (7): 1442–5. doi:10.1016/j.cell.2011.12.006. PMC 3254670. PMID 22196723.
- Grønlien HK, Berg T, Løvlie AM (July 2002). "In the polymorphic ciliate Tetrahymena vorax, the non-selective phagocytosis seen in microstomes changes to a highly selective process in macrostomes". The Journal of Experimental Biology. 205 (Pt 14): 2089–97. PMID 12089212.
- Montagnes, Djs; Barbosa, Ab; Boenigk, J; Davidson, K; Jürgens, K; Macek, M; Parry, Jd; Roberts, Ec; imek, K (2008-09-18). "Selective feeding behaviour of key free-living protists: avenues for continued study". Aquatic Microbial Ecology. 53: 83–98. doi:10.3354/ame01229. ISSN 0948-3055.
- Stibor H, Sommer U (April 2003). "Mixotrophy of a photosynthetic flagellate viewed from an optimal foraging perspective". Protist. 154 (1): 91–8. doi:10.1078/143446103764928512. PMID 12812372.
External links

- Phagocytosis at the US National Library of Medicine Medical Subject Headings (MeSH)