Paxillin
Paxillin is a protein that in humans is encoded by the PXN gene. Paxillin is expressed at focal adhesions of non-striated cells and at costameres of striated muscle cells, and it functions to adhere cells to the extracellular matrix. Mutations in PXN as well as abnormal expression of paxillin protein has been implicated in the progression of various cancers.
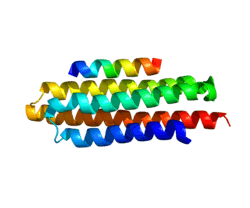
Structure
Human paxillin is 64.5 kDa in molecular weight and 591 amino acids in length.[5]
The C-terminal region of paxillin is composed of four tandem double zinc finger LIM domains that are cysteine/histidine-rich with conserved repeats; these serve as binding sites for the protein tyrosine phosphatase-PEST,[6] tubulin[7] and serves as the targeting motif for focal adhesions.[8]
The N-terminal region of paxillin has five highly conserved leucine-rich sequences termed LD motifs, which mediate several interactions, including that with pp125FAK and vinculin.[9][10] The LD motifs are predicted to form amphipathic alpha helices, with each leucine residue positioned on one face of the alpha helix to form a hydrophobic protein-binding interface. The N-terminal region also has a proline-rich domain that has potential for Src-SH3 binding. Three N-terminal YXXP motifs may serve as binding sites for talin or v-Crk SH2.[11][12]
Function
Paxillin is a signal transduction adaptor protein discovered in 1990 in the laboratory of Keith Burridge[13] The C-terminal region of paxillin contains four LIM domains that target paxillin to focal adhesions. It is presumed through a direct association with the cytoplasmic tail of beta-integrin. The N-terminal region of paxillin is rich in protein–protein interaction sites. The proteins that bind to paxillin are diverse and include protein tyrosine kinases, such as Src and focal adhesion kinase (FAK), structural proteins, such as vinculin and actopaxin, and regulators of actin organization, such as COOL/PIX and PKL/GIT. Paxillin is tyrosine-phosphorylated by FAK and Src upon integrin engagement or growth factor stimulation,[14] creating binding sites for the adapter protein Crk.
In striated muscle cells, paxillin is important in costamerogenesis, or the formation of costameres, which are specialized focal adhesion-like structures in muscle cells that tether Z-disc structures across the sarcolemma to the extracellular matrix. The current working model of costamerogenesis is that in cultured, undifferentiated myoblasts, alpha-5 integrin, vinculin and paxillin are in complex and located primarily at focal adhesions. During early differentiation, premyofibril formation through sarcomerogenesis occurs, and premyofibrils assemble at structures that are typical of focal adhesions in non-muscle cells; a similar phenomenon is observed in cultured cardiomyocytes.[15] Premyofibrils become nascent myofibrils, which progressively align to form mature myofibrils and nascent costamere structures appear. Costameric proteins redistribute to form mature costameres.[16] While the precise functions of paxillin in this process are still being unveiled, studies investigating binding partners of paxillin have provided mechanistic understanding of its function. The proline-rich region of paxillin specifically binds to the second SH3 domain of ponsin, which occurs after the onset of the myogenic differentiation and with expression restricted to costameres.[17] We also know that the binding of paxillin to focal adhesion kinase (FAK) is critical for directing paxillin function. The phosphorylation of FAK at serine-910 regulates the interaction of FAK with paxillin, and controls the stability of paxillin at costameres in cardiomyocytes, with phosphorylation reducing the half-life of paxillin.[18] This is important to understand because the stability of the FAK-paxillin interaction is likely inversely related to the stability of the vinculin-paxillin interaction, which would likely indicate the strength of the costamere interaction as well as sarcomere reorganization; processes which have been linked to dilated cardiomyopathy.[19] Additional studies have shown that paxillin itself is phosphorylated, and this participates in hypertrophic signaling pathways in cardiomyocytes. Treatment of cardiomyocytes with the hypertrophic agonist, phenylephrine stimulated a rapid increase in tyrosine phosphorylation paxillin, which was mediated by protein tyrosine kinases.[20]
The structural reorganization of paxillin in cardiomyocytes has also been detected in mouse models of dilated cardiomyopathy. In a mouse model of tropomodulin overexpression, paxillin distribution was revamped coordinate with increased phosphorylation and cleavage of paxillin.[21] Similarly, paxillin was shown to have altered localization in cardiomyocytes from transgenic mice expressing a constitutively-active rac1.[22] These data show that alterations in costameric organization, in part via paxillin redistribution, may be a pathogenic mechanism in dilated cardiomyopathy. In addition, in mice subjected to pressure overload-induced cardiac hypertrophy, inducing hypertrophic cardiomyopathy, paxillin expression levels increased, suggesting a role for paxillin in both types of cardiomyopathy.[23]
Clinical significance
Paxillin has been shown to have a clinically-significant role in patients with several cancer types. Enhanced expression of paxillin has been detected in premalignant areas of hyperplasia, squamous metaplasia and goblet cell metaplasia, as well as dysplastic lesions and carcinoma in high-risk patients with lung adenocarcinoma.[24] Mutations in PXN have been associated with enhanced tumor growth, cell proliferation, and invasion in lung cancer tissues.[25]
During tumor transformation, a consistent finding is that paxillin protein is recruited and phosphorylated.[26] Paxillin plays a role in the MET tyrosine kinase signaling pathway, which is upregulated in many cancers.[27]
Interactions
Paxillin has been shown to interact with:
- PTP-PEST,[6]
- tubulin,[7]
- VCL,[13][28]
- pp125FAK.[29][30][31]
- SRC[32][33]
- SORBS1.[17]
- PARVA[34] and
- ILK[35]
References
- GRCh38: Ensembl release 89: ENSG00000089159 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000029528 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Protein sequence of human PXN (Uniprot ID: P49023)". Cardiac Organellar Protein Atlas Knowledgebase (COPaKB). Archived from the original on July 13, 2015. Retrieved July 13, 2015.
- Shen Y, Schneider G, Cloutier JF, Veillette A, Schaller MD (March 1998). "Direct association of protein-tyrosine phosphatase PTP-PEST with paxillin". The Journal of Biological Chemistry. 273 (11): 6474–81. doi:10.1074/jbc.273.11.6474. PMID 9497381.
- Herreros L, Rodríguez-Fernandez JL, Brown MC, Alonso-Lebrero JL, Cabañas C, Sánchez-Madrid F, Longo N, Turner CE, Sánchez-Mateos P (August 2000). "Paxillin localizes to the lymphocyte microtubule organizing center and associates with the microtubule cytoskeleton" (PDF). The Journal of Biological Chemistry. 275 (34): 26436–40. doi:10.1074/jbc.M003970200. PMID 10840040.
- Côté JF, Turner CE, Tremblay ML (July 1999). "Intact LIM 3 and LIM 4 domains of paxillin are required for the association to a novel polyproline region (Pro 2) of protein-tyrosine phosphatase-PEST". The Journal of Biological Chemistry. 274 (29): 20550–60. doi:10.1074/jbc.274.29.20550. PMID 10400685.
- Brown MC, Curtis MS, Turner CE (August 1998). "Paxillin LD motifs may define a new family of protein recognition domains". Nature Structural Biology. 5 (8): 677–8. doi:10.1038/1370. PMID 9699628.
- Tumbarello DA, Brown MC, Turner CE (February 2002). "The paxillin LD motifs". FEBS Letters. 513 (1): 114–8. doi:10.1016/s0014-5793(01)03244-6. PMID 11911889.
- Salgia R, Li JL, Lo SH, Brunkhorst B, Kansas GS, Sobhany ES, Sun Y, Pisick E, Hallek M, Ernst T (March 1995). "Molecular cloning of human paxillin, a focal adhesion protein phosphorylated by P210BCR/ABL". The Journal of Biological Chemistry. 270 (10): 5039–47. doi:10.1074/jbc.270.10.5039. PMID 7534286.
- Turner CE (September 1998). "Paxillin". The International Journal of Biochemistry & Cell Biology. 30 (9): 955–9. doi:10.1016/s1357-2725(98)00062-4. PMID 9785458.
- Turner CE, Glenney JR, Burridge K (1990). "Paxillin: a new vinculin-binding protein present in focal adhesions". J. Cell Biol. 111 (3): 1059–68. doi:10.1083/jcb.111.3.1059. PMC 2116264. PMID 2118142.
- Bellis SL, Miller JT, Turner CE (July 1995). "Characterization of tyrosine phosphorylation of paxillin in vitro by focal adhesion kinase". The Journal of Biological Chemistry. 270 (29): 17437–41. doi:10.1074/jbc.270.29.17437. PMID 7615549.
- Decker ML, Simpson DG, Behnke M, Cook MG, Decker RS (July 1990). "Morphological analysis of contracting and quiescent adult rabbit cardiac myocytes in long-term culture". The Anatomical Record. 227 (3): 285–99. doi:10.1002/ar.1092270303. PMID 2372136.
- Quach NL, Rando TA (May 2006). "Focal adhesion kinase is essential for costamerogenesis in cultured skeletal muscle cells". Developmental Biology. 293 (1): 38–52. doi:10.1016/j.ydbio.2005.12.040. PMID 16533505.
- Gehmlich K, Pinotsis N, Hayess K, van der Ven PF, Milting H, El Banayosy A, Körfer R, Wilmanns M, Ehler E, Fürst DO (June 2007). "Paxillin and ponsin interact in nascent costameres of muscle cells". Journal of Molecular Biology. 369 (3): 665–82. doi:10.1016/j.jmb.2007.03.050. PMID 17462669.
- Chu M, Iyengar R, Koshman YE, Kim T, Russell B, Martin JL, Heroux AL, Robia SL, Samarel AM (December 2011). "Serine-910 phosphorylation of focal adhesion kinase is critical for sarcomere reorganization in cardiomyocyte hypertrophy". Cardiovascular Research. 92 (3): 409–19. doi:10.1093/cvr/cvr247. PMC 3246880. PMID 21937583.
- Zemljic-Harpf AE, Miller JC, Henderson SA, Wright AT, Manso AM, Elsherif L, Dalton ND, Thor AK, Perkins GA, McCulloch AD, Ross RS (November 2007). "Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy". Molecular and Cellular Biology. 27 (21): 7522–37. doi:10.1128/MCB.00728-07. PMC 2169049. PMID 17785437.
- Taylor JM, Rovin JD, Parsons JT (June 2000). "A role for focal adhesion kinase in phenylephrine-induced hypertrophy of rat ventricular cardiomyocytes". The Journal of Biological Chemistry. 275 (25): 19250–7. doi:10.1074/jbc.M909099199. PMID 10749882.
- Melendez J, Welch S, Schaefer E, Moravec CS, Avraham S, Avraham H, Sussman MA (November 2002). "Activation of pyk2/related focal adhesion tyrosine kinase and focal adhesion kinase in cardiac remodeling". The Journal of Biological Chemistry. 277 (47): 45203–10. doi:10.1074/jbc.M204886200. PMID 12228222.
- Sussman MA, Welch S, Walker A, Klevitsky R, Hewett TE, Price RL, Schaefer E, Yager K (April 2000). "Altered focal adhesion regulation correlates with cardiomyopathy in mice expressing constitutively active rac1". The Journal of Clinical Investigation. 105 (7): 875–86. doi:10.1172/JCI8497. PMC 377478. PMID 10749567.
- Yund EE, Hill JA, Keller RS (October 2009). "Hic-5 is required for fetal gene expression and cytoskeletal organization of neonatal cardiac myocytes". Journal of Molecular and Cellular Cardiology. 47 (4): 520–7. doi:10.1016/j.yjmcc.2009.06.006. PMC 3427732. PMID 19540241.
- Mackinnon AC, Tretiakova M, Henderson L, Mehta RG, Yan BC, Joseph L, Krausz T, Husain AN, Reid ME, Salgia R (January 2011). "Paxillin expression and amplification in early lung lesions of high-risk patients, lung adenocarcinoma and metastatic disease". Journal of Clinical Pathology. 64 (1): 16–24. doi:10.1136/jcp.2010.075853. PMC 3002839. PMID 21045234.
- Jagadeeswaran R, Surawska H, Krishnaswamy S, Janamanchi V, Mackinnon AC, Seiwert TY, Loganathan S, Kanteti R, Reichman T, Nallasura V, Schwartz S, Faoro L, Wang YC, Girard L, Tretiakova MS, Ahmed S, Zumba O, Soulii L, Bindokas VP, Szeto LL, Gordon GJ, Bueno R, Sugarbaker D, Lingen MW, Sattler M, Krausz T, Vigneswaran W, Natarajan V, Minna J, Vokes EE, Ferguson MK, Husain AN, Salgia R (January 2008). "Paxillin is a target for somatic mutations in lung cancer: implications for cell growth and invasion". Cancer Research. 68 (1): 132–42. doi:10.1158/0008-5472.CAN-07-1998. PMC 2767335. PMID 18172305.
- Vande Pol SB, Brown MC, Turner CE (January 1998). "Association of Bovine Papillomavirus Type 1 E6 oncoprotein with the focal adhesion protein paxillin through a conserved protein interaction motif". Oncogene. 16 (1): 43–52. doi:10.1038/sj.onc.1201504. PMID 9467941.
- Lawrence RE, Salgia R (2010). "MET molecular mechanisms and therapies in lung cancer". Cell Adhesion & Migration. 4 (1): 146–52. doi:10.4161/cam.4.1.10973. PMC 2852571. PMID 20139696.
- Wood CK, Turner CE, Jackson P, Critchley DR (February 1994). "Characterisation of the paxillin-binding site and the C-terminal focal adhesion targeting sequence in vinculin". Journal of Cell Science. 107 (2): 709–17. PMID 8207093.
- Turner CE, Miller JT (June 1994). "Primary sequence of paxillin contains putative SH2 and SH3 domain binding motifs and multiple LIM domains: identification of a vinculin and pp125Fak-binding region". Journal of Cell Science. 107 (6): 1583–91. PMID 7525621.
- Hildebrand JD, Schaller MD, Parsons JT (June 1995). "Paxillin, a tyrosine phosphorylated focal adhesion-associated protein binds to the carboxyl terminal domain of focal adhesion kinase". Molecular Biology of the Cell. 6 (6): 637–47. doi:10.1091/mbc.6.6.637. PMC 301225. PMID 7579684.
- Brown MC, Perrotta JA, Turner CE (November 1996). "Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding". The Journal of Cell Biology. 135 (4): 1109–23. doi:10.1083/jcb.135.4.1109. PMC 2133378. PMID 8922390.
- Turner CE (December 2000). "Paxillin interactions". Journal of Cell Science. 113 (23): 4139–40. PMID 11069756.
- Turner CE (December 2000). "Paxillin and focal adhesion signalling". Nature Cell Biology. 2 (12): E231-6. doi:10.1038/35046659. PMID 11146675.
- Nikolopoulos SN, Turner CE (December 2000). "Actopaxin, a new focal adhesion protein that binds paxillin LD motifs and actin and regulates cell adhesion". The Journal of Cell Biology. 151 (7): 1435–48. doi:10.1083/jcb.151.7.1435. PMC 2150668. PMID 11134073.
- Nikolopoulos SN, Turner CE (June 2001). "Integrin-linked kinase (ILK) binding to paxillin LD1 motif regulates ILK localization to focal adhesions". The Journal of Biological Chemistry. 276 (26): 23499–505. doi:10.1074/jbc.M102163200. PMID 11304546.
Further reading
- Panetti TS (January 2002). "Tyrosine phosphorylation of paxillin, FAK, and p130CAS: effects on cell spreading and migration". Frontiers in Bioscience. 7 (1–3): d143-50. doi:10.2741/panetti. PMID 11779709.
- Rose DM, Han J, Ginsberg MH (August 2002). "Alpha4 integrins and the immune response". Immunological Reviews. 186: 118–24. doi:10.1034/j.1600-065X.2002.18611.x. PMID 12234367.
- Salgia R, Uemura N, Okuda K, Li JL, Pisick E, Sattler M, de Jong R, Druker B, Heisterkamp N, Chen LB (December 1995). "CRKL links p210BCR/ABL with paxillin in chronic myelogenous leukemia cells". The Journal of Biological Chemistry. 270 (49): 29145–50. doi:10.1074/jbc.270.49.29145. PMID 7493940.
- Bergman M, Joukov V, Virtanen I, Alitalo K (February 1995). "Overexpressed Csk tyrosine kinase is localized in focal adhesions, causes reorganization of alpha v beta 5 integrin, and interferes with HeLa cell spreading". Molecular and Cellular Biology. 15 (2): 711–22. doi:10.1128/mcb.15.2.711. PMC 231937. PMID 7529872.
- Schaller MD, Otey CA, Hildebrand JD, Parsons JT (September 1995). "Focal adhesion kinase and paxillin bind to peptides mimicking beta integrin cytoplasmic domains". The Journal of Cell Biology. 130 (5): 1181–7. doi:10.1083/jcb.130.5.1181. PMC 2120552. PMID 7657702.
- Yoshida M, Westlin WF, Wang N, Ingber DE, Rosenzweig A, Resnick N, Gimbrone MA (April 1996). "Leukocyte adhesion to vascular endothelium induces E-selectin linkage to the actin cytoskeleton". The Journal of Cell Biology. 133 (2): 445–55. doi:10.1083/jcb.133.2.445. PMC 2120789. PMID 8609175.
- Salgia R, Sattler M, Pisick E, Li JL, Griffin JD (February 1996). "p210BCR/ABL induces formation of complexes containing focal adhesion proteins and the protooncogene product p120c-Cbl". Experimental Hematology. 24 (2): 310–3. PMID 8641358.
- Salgia R, Pisick E, Sattler M, Li JL, Uemura N, Wong WK, Burky SA, Hirai H, Chen LB, Griffin JD (October 1996). "p130CAS forms a signaling complex with the adapter protein CRKL in hematopoietic cells transformed by the BCR/ABL oncogene". The Journal of Biological Chemistry. 271 (41): 25198–203. doi:10.1074/jbc.271.41.25198. PMID 8810278.
- Retta SF, Barry ST, Critchley DR, Defilippi P, Silengo L, Tarone G (December 1996). "Focal adhesion and stress fiber formation is regulated by tyrosine phosphatase activity". Experimental Cell Research. 229 (2): 307–17. doi:10.1006/excr.1996.0376. hdl:2318/37568. PMID 8986614.
- Mazaki Y, Hashimoto S, Sabe H (March 1997). "Monocyte cells and cancer cells express novel paxillin isoforms with different binding properties to focal adhesion proteins". The Journal of Biological Chemistry. 272 (11): 7437–44. doi:10.1074/jbc.272.11.7437. PMID 9054445.
- Hiregowdara D, Avraham H, Fu Y, London R, Avraham S (April 1997). "Tyrosine phosphorylation of the related adhesion focal tyrosine kinase in megakaryocytes upon stem cell factor and phorbol myristate acetate stimulation and its association with paxillin". The Journal of Biological Chemistry. 272 (16): 10804–10. doi:10.1074/jbc.272.16.10804. PMID 9099734.
- Ostergaard HL, Lou O, Arendt CW, Berg NN (March 1998). "Paxillin phosphorylation and association with Lck and Pyk2 in anti-CD3- or anti-CD45-stimulated T cells". The Journal of Biological Chemistry. 273 (10): 5692–6. doi:10.1074/jbc.273.10.5692. PMID 9488700.
- Fernandez R, Suchard SJ (May 1998). "Syk activation is required for spreading and H2O2 release in adherent human neutrophils". Journal of Immunology. 160 (10): 5154–62. PMID 9590268.
- Lewis JM, Schwartz MA (June 1998). "Integrins regulate the association and phosphorylation of paxillin by c-Abl". The Journal of Biological Chemistry. 273 (23): 14225–30. doi:10.1074/jbc.273.23.14225. PMID 9603926.
- Ganju RK, Munshi N, Nair BC, Liu ZY, Gill P, Groopman JE (July 1998). "Human immunodeficiency virus tat modulates the Flk-1/KDR receptor, mitogen-activated protein kinases, and components of focal adhesion in Kaposi's sarcoma cells". Journal of Virology. 72 (7): 6131–7. PMC 110419. PMID 9621077.
- Deakin NO, Turner CE (August 2008). "Paxillin comes of age". Journal of Cell Science. 121 (Pt 15): 2435–44. doi:10.1242/jcs.018044. PMC 2522309. PMID 18650496.
External links
- MBInfo: Paxillin
- Paxillin Info with links in the Cell Migration Gateway
- Paxillin at the US National Library of Medicine Medical Subject Headings (MeSH)
- Overview of all the structural information available in the PDB for UniProt: P49023 (Human Paxillin) at the PDBe-KB.
- Overview of all the structural information available in the PDB for UniProt: Q8VI36 (Mouse Paxillin) at the PDBe-KB.