NiFe hydrogenase
[NiFe] hydrogenase is a type of hydrogenase, which is an oxidative enzyme that reversibly convert molecular hydrogen in prokaryotes including Bacteria and Archaea.[1][2] The catalytic site on the enzyme provides simple hydrogen-metabolizing microorganisms a redox mechanism by which to store and utilize energy via the reaction shown in Figure 1. This is particularly essential for the anaerobic, sulfate-reducing bacteria of the genus Desulfovibrio[3][4] as well as pathogenic organisms Escherichia coli and Helicobacter pylori.[2] The mechanisms, maturation, and function of [NiFe] hydrogenases are actively being researched for applications to the hydrogen economy and as potential antibiotic targets.
Structure
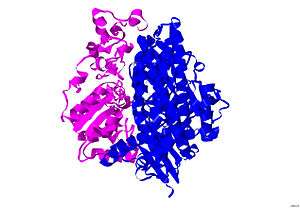
The structure of [NiFe] hydrogenase was obtained from x-ray crystallography studies of five different sulfate-reducing bacteria: D. vulgaris Miyazaki F,[6] D. gigas,[7] D. frutosovorans,[8][9] D. desulfuricans[10] and Desulfomicrobium baculatum.[11] The [NiFe] hydrogenase isolated from D. vulgaris Miyazaki F is shown in Figure 2. The larger subunit is in blue, has a molecular mass of 62.5 kDa, and houses the Ni-Fe active site. The smaller subunit is in magenta, has a molecular mass of 28.8 kDa, and contains the Fe-S clusters (see Iron-sulfur cluster).
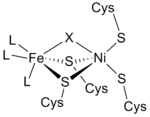
From the infrared spectra and X-ray crystallography studies, the [NiFe] hydrogenase active site was found to be (S-Cys)4Ni(μ-X)Fe(CO)(CN)2, in which the generic ligand X is either an oxide, sulfur, hydroperoxide, or a hydroxide found in an oxidized state only (Figure 3).[12] While the nickel atom participates in redox reactions, the iron atom is consistently in a Fe(II) coordination state.[12] The exact geometry of the three non-protein ligands (denoted as L) coordinating to the Fe metal ion is not known; however, they were identified as one carbon monoxide (C≡O) molecule and two cyanide (−C≡N) molecules.[13]
Fe-S clusters
Almost all hydrogenases contain at least one iron-sulfur cluster (Fe-S cluster). As previously mentioned, these Fe-S clusters connect the nickel active site of the enzyme to the surface of the protein because they serve as an electron transport chain from the Ni-Fe redox site to the electron acceptor cytochrome c3 (see Cytochrome c family).[13] These electrons are produced from the heterolytic cleavage of the hydrogen molecule at the Ni-Fe active site. Crystal structures of the hydrogenase show a Fe3S4 in the center of the chain, and a Fe4S4 cluster at the molecular surface. The distance between the internal Fe4S4 cluster and the active site is approximately 12 Å (Figure 4).[13]
The [NiFe] and [NiFeSe] hydrogenases have remarkably similar structures, leading to the suggestion that one sulfur on a Fe-S cluster was replaced by a selenium atom, but these hydrogenases differ in catalytic reactivity and sensitivity to enzyme inhibitors.[4]

Mg metal ion and the proton pathways
[NiFe] hydrogenase has a Mg2+ cation bonded in the C-terminus region of the larger subunit. This cation is bonded to three water molecules and three amino acids, and it stabilizes this solvent-free region. At approximately 13 Å away from the [NiFe] moiety, this cation connects the active site to a hydrogen bonding network and serves as a proton (H+) transfer pathway.[13]
The gas-access channel
Studies in which xenon was bound to the hydrogenase suggest a hydrophobic gas channel through which H2, CO, and O2 gases could reach the deeply buried active site within the enzyme. Crystal structure revealed several small channels at the surface, which combined into one larger channel that reached the [Ni-Fe] active site.[13]
Since hydrogenases are well-known to be oxygen sensitive, the diffusion of gas to the active site depends on the size and environment of the gas-access channel, the reaction of molecular oxygen (O2) at the active site, and the recovery of the active site after oxidation.[13]
Mechanism
The exact reaction mechanism of [NiFe] hydrogenases has been a matter of great debate. In 2009, a mechanism was proposed by Higuchi and coworkers based on X-ray crystallography and spectroscopic data of Desulfovibrio vulgaris Miyazaki F.[13] During the catalytic process, the Fe metal ion in the active site does not change its oxidation state while the Ni metal ion participates in redox chemistry. There are two main groups of redox states that [NiFe] hydrogenases pass through during catalysis 1) Inactive redox states and 2) Active redox states (Figure 5).
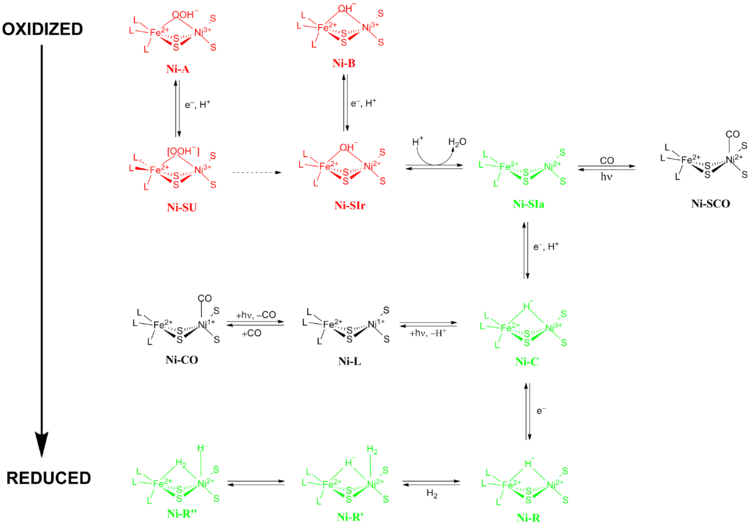
Inactive redox states
Ni-A (the “unready” state) and Ni-B (the “ready” state) are the most oxidized forms of the [NiFe] metal center and are activated via one-electron reduction with proton transfer. The rate of reductive activation of Ni-A to Ni-SU can take hours while the rate of reductive activation of Ni-B to Ni-SIr happens in seconds.[14] The reason for this disparity in activation kinetics between Ni-A and Ni-B was proposed to be a result of the difference in bridging ligands between the two different redox states. At the Ni-SIr state, a water molecule was released to form the Ni-SIa state, the first catalytic redox active state of [NiFe] hydrogenases.
Active redox states
The three most important catalytic redox active states of [NiFe] hydrogenases are Ni-SIa, Ni-C and Ni-R (which have three different variations:).[13] The light-sensitive Ni-C state can be obtained via one electron reduction of Ni-SIa. The electron paramagnetic resonance spectroscopic studies of the Ni-C state, which contained a Ni3+ with S=1/2 (see Spin-1/2) and a hydride bridging the two metals Ni and Fe, showed that the heterolytic cleavage of H2 takes place in the [NiFe] hydrogenase active site.
The CO-inhibited states
Ni-SIa state can be inhibited by CO, which binds directly to Ni metal ion in a bent conformation to form Ni-SCO (see below).[15] Since Ni-C is light sensitive, upon illumination at 100K results in Ni-L redox state. In the presence of CO, Ni-L forms Ni-CO state.
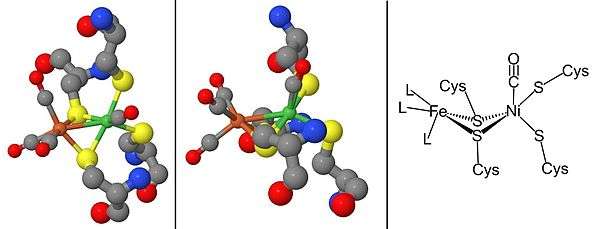
Maturation and genetic arrangement
The maturation of [NiFe] hydrogenases requires a set of accessory proteins that synthesize the NiFe active site, Figure 2, and modify the precursor enzyme so that it has the correct structure and location.[2][16][17] The maturation of the active site is of special interest because of the synthesis of cyanide (CN) and carbon monoxide (CO) metal ligands which are usually toxic to living organism.[16] This step is completed by the proteins HypC, HypD, HypE, and HypF.[17][18] After, synthesis of the iron center, nickel is inserted using metallochaperones HypA, HypB, and SlyD.[17][18] Once the catalytic center is completed, the hydrogenase precursor undergoes a C-terminal cleavage that prompts rearrangement of its structure and association with the small subunit.[16][17][18] Finally, the completed enzyme is transported to its correct position within the cell.[16][17][18] Hydrogenase promoter, PSH, can be studied constructing a PSH promoter-gfp fusion by using green fluorescent protein (gfp) reporter gene.[19]
Application
Since [NiFe] hydrogenase is a member of the hydrogenase family, these enzymes can catalyze both the consumption and production of hydrogen.[1] By studying [NiFe] hydrogenase, scientists can optimize a condition in which the protein will only produce hydrogen. Additionally, small enzyme mimic of [NiFe] hydrogenase can also be synthesized to act as hydrogen gas generator. The soluble [NiFe] hydrogenase from Ralstonia eutropha H16 is a promising candidate enzyme for H2-based biofuel application as it favours H2 oxidation and is relatively oxygen-tolerant. It can be produced on heterotrophic growth media [20] and purified via anion exchangeand size exclusion chromatography matrices.[21]
See also
- Hydrogenase
- Fe-Ni Clusters
- Bioinorganic
References
- Jugder, Bat-Erdene; Welch, Jeffrey; Aguey-Zinsou, Kondo-Francois; Marquis, Christopher P. (2013-05-14). "Fundamentals and electrochemical applications of [Ni–Fe]-uptake hydrogenases". RSC Advances. 3 (22): 8142. doi:10.1039/c3ra22668a. ISSN 2046-2069.
- Vignais, Paulette M.; Billoud, Bernard (October 2007). "Occurrence, Classification, and Biological Function of Hydrogenases: An Overview". Chemical Reviews. 107 (10): 4206–4272. doi:10.1021/cr050196r. PMID 17927159.
- Volbeda, A.; Garcin, E.; Piras, C.; de Lacey, A. L.; Fernandez, V. M.; Hatchikian, E. C.; Frey, M.; Fontecilla-Camps, J. C. (1996). "Structure of the [NiFe] Hydrogenase Active Site: Evidence for Biologically Uncommon Fe Ligands". J. Am. Chem. Soc. 118 (51): 12989–12996. doi:10.1021/ja962270g.
- Eidsness, M. K.; Scott, R. A.; Prickril, B. C.; DerVartanian, D. V.; Legall, J.; Moura, I.; Moura, J. J.; Peck, H. D. (1989). "Evidence for selenocysteine coordination to the active site nickel in the [NiFeSe]hydrogenases from Desulfovibrio baculatus". Proceedings of the National Academy of Sciences. 86 (1): 147–151. doi:10.1073/pnas.86.1.147. PMC 286421. PMID 2521386.
- Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
- Higuchi, Y.; Yagi, T.; Yasuoka, N. (1997). "Unusual ligand structure in Ni-Fe active center and an additional Mg site in hydrogenase revealed by high resolution X-ray structure analysis". Structure. 5 (12): 1671–1680. doi:10.1016/s0969-2126(97)00313-4. PMID 9438867.
- Volbeda, A.; Charon, M.-H.; Piras, C.; Hatchikian, E. C.; Frey, M.; Fontecilla-Camps, J. C. (1995). "Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas". Nature. 373 (6515): 580–587. doi:10.1038/373580a0. PMID 7854413.
- Volbeda, A.; Martin, L.; Cavazza, C.; Matho, M.; Faber, B. W.; Roseboom, W.; Albracht, S. P. J.; Garcin, E.; Rousset, M.; Fontecilla-Camps, J. C. (2005). "Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases". Journal of Biological Inorganic Chemistry. 10 (3): 239–249. doi:10.1007/s00775-005-0632-x. PMID 15803334.
- Montet, Y.; Amara, P.; Volbeda, A.; Vernede, X.; Hatchikian E. C.; Field, M. J.; Frey, M.; Fontecilla-Camps, J. C. (1997). "Gas access to the active site of Ni-Fe hydrogenases probed by X-ray crystallography and molecular dynamics". Nature Structural & Molecular Biology. 4 (7): 523–526. doi:10.1038/nsb0797-523. PMID 9228943.
- Matias, P. M.; Soares, C. M.; Saraiva, L. M.; Coelho, R.; Morais, J.; Le Gall, J.; Carrondo, M. A. (2001). "[NiFe] hydrogenase from Desulfovibrio desulfuricans ATCC 27774: gene sequencing, three-dimensional structure determination and refinement at 1.8 Å and modelling studies of its interaction with the tetrahaem cytochrome c3". Journal of Biological Inorganic Chemistry. 6 (1): 63–81. doi:10.1007/s007750000167. PMID 11191224.
- Garcin, E.; Vernede, X.; Hatchikian, E. C.; Volbeda, A.; Frey, M.; Fontecilla-Camps, J. C. (1999). "The crystal structure of a reduced [NiFeSe] hydrogenase provides an image of the activated catalytic center". Structure. 7 (5): 557–566. doi:10.1016/s0969-2126(99)80072-0. PMID 10378275.
- Chiou, T.-W.; Liaw, W.-F. (2008). "Nickel–thiolate and iron–thiolate cyanocarbonyl complexes: Modeling the nickel and iron sites of [NiFe] hydrogenase". Comptes Rendus Chimie. 11 (8): 818–833. doi:10.1016/j.crci.2008.04.003.
- Ogata, H.; Lubitz, W.; Higuchi, Y. (2009). "[NiFe] hydrogenases: structural and spectroscopic studies of the reaction mechanism". Dalton Trans. 37 (37): 7577–7587. doi:10.1039/b903840j. PMID 19759926.
- Lamele, S. E.; Albracht, S. P. J.; Armstrong, F. A. (2004). "Electrochemical Potential-Step Investigations of the Aerobic Interconversions of [NiFe]-Hydrogenase from Allochromatium vinosum: Insights into the Puzzling Difference between Unready and Ready Oxidized Inactive States". Journal of the American Chemical Society. 126 (45): 14899–14909. doi:10.1021/ja047939v. PMID 15535717.
- Ogata, H.; Mizoguchi, Y.; Mizuno, N.; Miki, K.; Adachi, S.-i.; Yasuoka, N.; Yagi, T.; Yamauchi, O.; Hirota, S.; Higuchi, Y. (2002). "Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen". Journal of the American Chemical Society. 124 (39): 11628–11635. doi:10.1021/ja012645k. PMID 12296727.
- Lubitz, Wolfgang; Ogata, Hideaki; Rüdiger, Olaf; Reijerse, Edward (23 April 2014). "Hydrogenases". Chemical Reviews. 114 (8): 4081–4148. doi:10.1021/cr4005814. PMID 24655035.
- Lacasse, Michael J.; Zamble, Deborah B. (29 March 2016). "[NiFe]-Hydrogenase Maturation". Biochemistry. 55 (12): 1689–1701. doi:10.1021/acs.biochem.5b01328. PMID 26919691.
- Peters, John W.; Schut, Gerrit J.; Boyd, Eric S.; Mulder, David W.; Shepard, Eric M.; Broderick, Joan B.; King, Paul W.; Adams, Michael W.W. (June 2015). "[FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1853 (6): 1350–1369. doi:10.1016/j.bbamcr.2014.11.021. PMID 25461840.
- Jugder, Bat-Erdene; Welch, Jeffrey; Braidy, Nady; Marquis, Christopher P. (2016-07-26). "Construction and use of aCupriavidus necatorH16 soluble hydrogenase promoter (PSH) fusion togfp(green fluorescent protein)". PeerJ. 4: e2269. doi:10.7717/peerj.2269. ISSN 2167-8359. PMC 4974937. PMID 27547572.
- Jugder, Bat-Erdene; Chen, Zhiliang; Ping, Darren Tan Tek; Lebhar, Helene; Welch, Jeffrey; Marquis, Christopher P. (2015-03-25). "An analysis of the changes in soluble hydrogenase and global gene expression in Cupriavidus necator ( Ralstonia eutropha ) H16 grown in heterotrophic diauxic batch culture". Microbial Cell Factories. 14 (1): 42. doi:10.1186/s12934-015-0226-4. ISSN 1475-2859. PMC 4377017. PMID 25880663.
- Jugder, Bat-Erdene; Lebhar, Helene; Aguey-Zinsou, Kondo-Francois; Marquis, Christopher P. (2016-01-01). "Production and purification of a soluble hydrogenase from Ralstonia eutropha H16 for potential hydrogen fuel cell applications". MethodsX. 3: 242–250. doi:10.1016/j.mex.2016.03.005. PMC 4816682. PMID 27077052.