Minicircle
Minicircles are small (~4kb) circular replicons. They occur naturally in some eukaryotic organelle genomes. In the mitochondria-derived kinetoplast of trypanosomes, minicircles encode guide RNAs for RNA editing.[1] In Amphidinium, the chloroplast genome is made of minicircles that encode chloroplast proteins.[2][3]
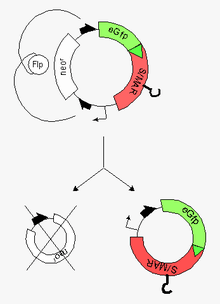
In vitro experimentally-derived minicircles
Minicircles are small (~4kb) circular plasmid derivatives that have been freed from all prokaryotic vector parts. They have been applied as transgene carriers for the genetic modification of mammalian cells, with the advantage that, since they contain no bacterial DNA sequences, they are less likely to be perceived as foreign and destroyed. (Typical transgene delivery methods involve plasmids, which contain foreign DNA.) The smaller size of minicircles also extends their cloning capacity and facilitates their delivery into cells.
Their preparation usually follows a two-step procedure:[4][5]
- production of a 'parental plasmid' (bacterial plasmid with eukaryotic inserts) in E. coli
- induction of a site-specific recombinase at the end of this process but still in bacteria. These steps are followed by the
- excision of prokaryotic vector parts via two recombinase-target sequences at both ends of the insert
- recovery of the resulting minicircle (vehicle for the highly efficient modification of the recipient cell) and the miniplasmid by capillary gel electrophoresis (CGE)
The purified minicircle can be transferred into the recipient cell by transfection or lipofection and into a differentiated tissue by, for instance, jet injection.
Conventional minicircles lack an origin of replication, so they do not replicate within the target cells and the encoded genes will disappear as the cell divides (which can be either an advantage or disadvantage depending on whether the application demands persistent or transient expression). A novel addition to the field are nonviral self-replicating minicircles, which owe this property to the presence of a S/MAR-Element. Self-replicating minicircles hold great promise for the systematic modification of stem cells and will significantly extend the potential of their plasmidal precursor forms ("parental plasmids"), the more as the principal feasibility of such an approach has amply been demonstrated for their plasmidal precursor forms.[6][7][8][9]
See also
References
- "Kinetoplastids and Their Networks of Interlocked DNA". Retrieved September 2, 2019.
- Barbrook, Adrian C.; Voolstra, Christian R.; Howe, Christopher J. (2014). "The Chloroplast Genome of a Symbiodinium sp. Clade C3 Isolate". Protist. 165 (1): 1–13. doi:10.1016/j.protis.2013.09.006. PMID 24316380.
- Dorrell, Richard G.; Nisbet, R. Ellen R.; Barbrook, Adrian C.; Rowden, Stephen J.L.; Howe, Christopher J. (2019). "Integrated Genomic and Transcriptomic Analysis of the Peridinin Dinoflagellate Amphidinium carterae Plastid". Protist. 170 (4): 358–373. doi:10.1016/j.protis.2019.06.001. PMID 31415953.
- Nehlsen, K., Broll S., Bode, J. (2006). "Replicating minicircles: Generation of nonviral episomes for the efficient modification of dividing cells". Gene Ther. Mol. Biol. 10: 233–244.
- Kay, M.A., He, C.-Y, Chen, Z.-H. (2010). "A robust system for production of minicircle DNA vectors". Nature Biotechnology. 28 (12): 1287–1289. doi:10.1038/nbt.1708. PMC 4144359. PMID 21102455.
- Broll, S., Oumard A., Hahn K., Schambach A, Bode, J. . (2010). "Minicircle Performance Depending on S/MAR-Nuclear Matrix Interactions". J. Mol. Biol. 395 (5): 950–965. doi:10.1016/j.jmb.2009.11.066. PMID 20004666.
- Argyros, O., Wong SP., Fedonidis C.; et al. (2011). "Development of S/MAR minicircles for enhanced and persistent transgene expression in the mouse liver". J. Mol. Med. 89 (5): 515–529. doi:10.1007/s00109-010-0713-3. PMID 21301798.
- Heinz, N, Broll S, Schleef M, Baum C, Bode J (2012). "Filling a gap: S/MAR-based replicating minicircles". CliniBook - Nonviral Platform; Clinigene Network: 271–277.
- Nehlsen, Kristina; Broll, Sandra; Kandimalla, Raju; Heinz, Niels; Heine, Markus; Binius, Stefanie; Schambach, Axel; Bode, Jürgen (5 April 2013). "Replicating Minicircles: Overcoming the Limitations of Transient and Stable Expression Systems". In Schleef, Martin (ed.). Minicircle and Miniplasmid DNA Vectors: The Future of Nonviral and Viral Gene Transfer. Wiley‐VCH Verlag GmbH & Co. KGaA. pp. 115–162. doi:10.1002/9783527670420.ch8. ISBN 9783527670420.