Methylobacterium
The Methylobacteria are a genus of Rhizobiales.[3]
| Methylobacterium | |
|---|---|
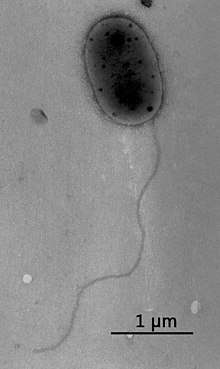 | |
| Scientific classification | |
| Kingdom: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | Methylobacterium Patt et al. 1976 |
| Type species | |
| Methylobacterium organophilum | |
| Species | |
|
M. adhaesivum | |
As well as its normal habitats in soil and water, Methylobacterium has also been identified as a contaminant of DNA extraction kit reagents, which may lead to its erroneous appearance in microbiota or metagenomic datasets.[4]
Natural genetic transformation
Natural genetic transformation in bacteria is a process involving transfer of DNA from one cell to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination. Methylobacterium organophilum cells are able to undergo genetic transformation and become competent for DNA uptake near the end of the exponential growth phase.[5]
References
- Parte, A.C. "Methylobacterium". LPSN.
- Madhaiyan, M; Poonguzhali, S (July 2014). "Methylobacterium pseudosasicola sp. nov. and Methylobacterium phyllostachyos sp. nov., isolated from bamboo leaf surfaces". International Journal of Systematic and Evolutionary Microbiology. 64 (Pt 7): 2376–84. doi:10.1099/ijs.0.057232-0. PMID 24760798.
- Garrity, George M.; Brenner, Don J.; Krieg, Noel R.; Staley, James T. (eds.) (2005). Bergey's Manual of Systematic Bacteriology, Volume Two: The Proteobacteria, Part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. New York, New York: Springer. ISBN 978-0-387-24145-6.
- Salter, S; Cox, M; Turek, E; Calus, S; Cookson, W; Moffatt, M; Turner, P; Parkhill, J; Loman, N; Walker, A (2014). "Reagent contamination can critically impact sequence-based microbiome analyses". bioRxiv 10.1101/007187.
- O'Connor M, Wopat A, Hanson RS (1977). "Genetic transformation in Methylobacterium organophilum". J. Gen. Microbiol. 98 (1): 265–72. doi:10.1099/00221287-98-1-265. PMID 401866.