C4 carbon fixation
C4 carbon fixation or the Hatch–Slack pathway is a photosynthetic process in some plants. It is the first step in extracting carbon from carbon dioxide (CO
2) to be able to use it in sugar and other biomolecules. It is one of three known processes for carbon fixation. C4 refers to the four-carbon molecule that is the first product of this type of carbon fixation.
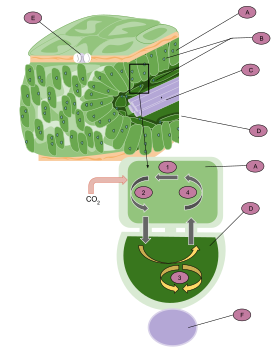
A: Mesophyll cell
B: Chloroplast
C: Vascular tissue
D: Bundle sheath cell
E: Stoma
F: Vascular tissue
1. CO
2 is fixed to produce a four-carbon molecule (malate or aspartate).
2. The molecule exits the cell and enters the bundle sheath cells.
3. It is then broken down into CO
2 and pyruvate. CO
2 enters the Calvin cycle to produce carbohydrates.
4. Pyruvate reenters the mesophyll cell, where it is reused to produce malate or aspartate.
C4 fixation is an elaboration of the more common C3 carbon fixation and is believed to have evolved more recently. C4 overcomes the tendency of the enzyme RuBisCO to wastefully fix oxygen rather than carbon dioxide in the process of photorespiration. This is achieved by ensuring that RuBisCO works in an environment where there is a lot of carbon dioxide and very little oxygen. CO
2 is shuttled via malate or aspartate from mesophyll cells to bundle-sheath cells. In these bundle-sheath cells CO
2 is released by decarboxylation of the malate. C4 plants use PEP carboxylase to capture more CO
2 in the mesophyll cells. PEP (phosphoenolpyruvate, three carbons) binds to CO
2 to make oxaloacetic acid (OAA, four carbons). OAA then makes malate or aspartate (four carbons), which is transported into the bundle sheath cells, where it releases the CO
2. These additional steps, however, require more energy in the form of ATP. Using this extra energy, C4 plants are able to more efficiently fix carbon in drought, high temperatures, and limitations of nitrogen or CO
2. Since the more common C3 pathway does not require this extra energy, it is more efficient in the other conditions.
The naming Hatch–Slack pathway is in honor of Marshall Davidson Hatch and Charles Roger Slack, who elucidated it in Australia in 1966.[1]
Discovery and resolution of the mechanism
The first experiments indicating that some plants do not use C3 carbon fixation but instead produce malate and aspartate in the first step of carbon fixation were done in the 1950s and early 1960s by Hugo Peter Kortschak and Yuri Karpilov.[2][3] The C4 pathway was elucidated by Marshall Davidson Hatch and Charles Roger Slack, in Australia, in 1966;[1] it is sometimes called the Hatch–Slack pathway.
C4 pathway
In C3 plants, the first step in the light-independent reactions of photosynthesis involves the fixation of CO
2 by the enzyme RuBisCO into 3-phosphoglycerate. However, due to the dual carboxylase and oxygenase activity of RuBisCo, some part of the substrate is oxidized rather than carboxylated, resulting in loss of substrate and consumption of energy, in what is known as photorespiration.
In order to bypass the photorespiration pathway, C4 plants have developed a mechanism to efficiently deliver CO
2 to the RuBisCO enzyme. They use their specific leaf anatomy where chloroplasts exist not only in the mesophyll cells in the outer part of their leaves but in the bundle sheath cells as well. Instead of direct fixation by RuBisCO into the three-carbon compound 3-phosphoglycerate, CO
2 is incorporated into a four-carbon organic acid (either malate or aspartate). The organic acid is produced in the mesophyll cells and then transported through plasmodesmata into the bundle sheath cells, where it is decarboxylated to regenerate CO
2. The chloroplasts of the bundle sheath cells can then use this CO
2 to produce carbohydrates by the conventional C3 pathway.
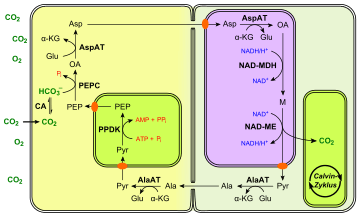
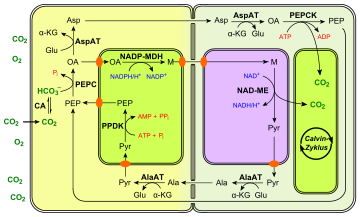
There are several variants of the C4 pathways, all of which use a four-carbon organic acid to transport CO
2 from the mesophyll cells to the bundle sheath cells. They achieve this purpose by using different substrates and enzymes. Three examples are provided in the figures on the right. The NADP-ME type C4 pathway is described in detail below.
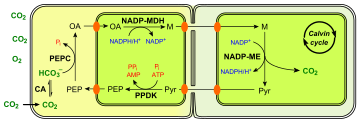
NADP-ME type
The first step in the NADP-ME type C4 pathway is the conversion of pyruvate (Pyr) to phosphoenolpyruvate (PEP), by the enzyme Pyruvate phosphate dikinase (PPDK). This reaction requires inorganic phosphate and ATP plus pyruvate, producing PEP, AMP, and inorganic pyrophosphate (PPi). The next step is the fixation of CO
2 into oxaloacetate by the PEP carboxylase enzyme (PEPC). Both of these steps occur in the mesophyll cells:
- pyruvate + Pi + ATP → PEP + AMP + PPi
- PEP + CO
2 → oxaloacetate
PEPC has a lower Km for HCO−
3 — and, hence, higher affinity — than RuBisCO. Furthermore, O2 is a very poor substrate for this enzyme. Thus, at relatively low concentrations of CO
2, most CO
2 will be fixed by this pathway.
The product is usually converted to malate (M), a simple organic compound, which is transported to the bundle-sheath cells surrounding a nearby vein. Here, it is decarboxylated by the NADP-malic enzyme (NADP-ME) to produce CO
2 and pyruvate. The CO
2 now enters the Calvin cycle and the pyruvate is transported back to the mesophyll cell.
Other types
There are several variants of this pathway, in which different substrates and enzymes are used to transport CO
2 from the mesophyll cells to the bundle sheath cells:
- The four-carbon acid transported from mesophyll cells may be malate (M), as above, or aspartate (Asp), or both (see the PEPCK type C4 pathway).
- The three-carbon acid transported back from bundle-sheath cells may be pyruvate (Pyr), as above, or alanine (Ala).
- The enzyme that catalyses decarboxylation in bundle-sheath cells differs:
- In maize and sugarcane, the enzyme is NADP-malic enzyme (NADP-ME);
- In millet, it is NAD-malic enzyme (NAD-ME);
- In megathyrsus maximus, it is PEP carboxykinase (PEPCK).
Efficiency
Since every CO
2 molecule has to be fixed twice, first in the mesphyll cells (by the PEPC enzyme) and second in the bundle sheath cells (by RuBisCO), the C4 pathway uses more energy than the C3 pathway. For instance, the C3 pathway requires 18 molecules of ATP for the synthesis of one molecule of glucose, whereas the NADP-ME type C4 pathway used by maize and sugarcane requires 30 molecules of ATP. This energy debt is more than paid for by avoiding losing more than half of photosynthetic carbon in photorespiration as occurs in some tropical plants, making it an adaptive mechanism for minimizing the loss.
Kranz leaf anatomy
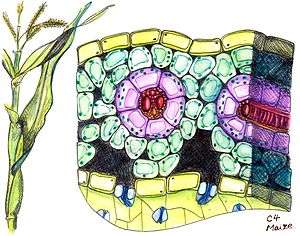
The C4 plants often possess a characteristic leaf anatomy called kranz anatomy, from the German word for wreath. Their vascular bundles are surrounded by two rings of cells; the inner ring, called bundle sheath cells, contains starch-rich chloroplasts lacking grana, which differ from those in mesophyll cells present as the outer ring. Hence, the chloroplasts are called dimorphic. The primary function of kranz anatomy is to provide a site in which CO
2 can be concentrated around RuBisCO, thereby avoiding photorespiration. In order to maintain a significantly higher CO
2 concentration in the bundle sheath compared to the mesophyll, the boundary layer of the kranz has a low conductance to CO
2, a property that may be enhanced by the presence of suberin.[4] The carbon concentration mechanism in C4 plants distinguishes their isotopic signature from other photosynthetic organisms.
Although most C4 plants exhibit kranz anatomy, there are, however, a few species that operate a limited C4 cycle without any distinct bundle sheath tissue. Suaeda aralocaspica, Bienertia cycloptera, Bienertia sinuspersici and Bienertia kavirense (all chenopods) are terrestrial plants that inhabit dry, salty depressions in the deserts of the Middle East. These plants have been shown to operate single-cell C4 CO
2-concentrating mechanisms, which are unique among the known C4 mechanisms.[5][6][7][8] Although the cytology of both genera differs slightly, the basic principle is that fluid-filled vacuoles are employed to divide the cell into two separate areas. Carboxylation enzymes in the cytosol can, therefore, be kept separate from decarboxylase enzymes and RuBisCO in the chloroplasts, and a diffusive barrier can be established between the chloroplasts (which contain RuBisCO) and the cytosol. This enables a bundle-sheath-type area and a mesophyll-type area to be established within a single cell. Although this does allow a limited C4 cycle to operate, it is relatively inefficient, with the occurrence of much leakage of CO
2 from around RuBisCO. There is also evidence for the exhibiting of inducible C4 photosynthesis by non-kranz aquatic macrophyte Hydrilla verticillata under warm conditions, although the mechanism by which CO
2 leakage from around RuBisCO is minimised is currently uncertain.[9]
Evolution and advantages
C4 plants have a competitive advantage over plants possessing the more common C3 carbon fixation pathway under conditions of drought, high temperatures, and nitrogen or CO
2 limitation. When grown in the same environment, at 30 °C, C3 grasses lose approximately 833 molecules of water per CO
2 molecule that is fixed, whereas C4 grasses lose only 277. This increased water use efficiency of C4 grasses means that soil moisture is conserved, allowing them to grow for longer in arid environments.[10]
C4 carbon fixation has evolved on up to 61 independent occasions in 19 different families of plants, making it a prime example of convergent evolution.[11] This convergence may have been facilitated by the fact that many potential evolutionary pathways to a C4 phenotype exist, many of which involve initial evolutionary steps not directly related to photosynthesis.[12] C4 plants arose around 35 million years ago[11] during the Oligocene (precisely when is difficult to determine) and did not become ecologically significant until around 6 to 7 million years ago, in the Miocene.[13] C4 metabolism in grasses originated when their habitat migrated from the shady forest undercanopy to more open environments,[14] where the high sunlight gave it an advantage over the C3 pathway.[15] Drought was not necessary for its innovation; rather, the increased resistance to water stress was a byproduct of the pathway and allowed C4 plants to more readily colonize arid environments.[15]
Today, C4 plants represent about 5% of Earth's plant biomass and 3% of its known plant species.[10][16] Despite this scarcity, they account for about 23% of terrestrial carbon fixation.[13][17] Increasing the proportion of C4 plants on earth could assist biosequestration of CO
2 and represent an important climate change avoidance strategy. Present-day C4 plants are concentrated in the tropics and subtropics (below latitudes of 45 degrees) where the high air temperature contributes to higher possible levels of oxygenase activity by RuBisCO, which increases rates of photorespiration in C3 plants.
Plants that use C4 carbon fixation
About 8,100 plant species use C4 carbon fixation, which represents about 3% of all terrestrial species of plants.[17][18] All these 8,100 species are angiosperms. C4 carbon fixation is more common in monocots compared with dicots, with 40% of monocots using the C4 pathway, compared with only 4.5% of dicots. Despite this, only three families of monocots use C4 carbon fixation compared to 15 dicot families. Of the monocot clades containing C4 plants, the grass (Poaceae) species use the C4 photosynthetic pathway most. 46% of grasses are C4 and together account for 61% of C4 species. C4 has arisen independently in the grass family some twenty or more times, in various subfamilies, tribes, and genera,[19] including the Andropogoneae tribe which contains the food crops maize, sugar cane, and sorghum. Various kinds of millet are also C4.[20][21] Of the dicot clades containing C4 species, the order Caryophyllales contains the most species. Of the families in the Caryophyllales, the Chenopodiaceae use C4 carbon fixation the most, with 550 out of 1,400 species using it. About 250 of the 1,000 species of the related Amaranthaceae also use C4.[10][22]
Members of the sedge family Cyperaceae, and members of numerous families of eudicots – including Asteraceae (the daisy family), Brassicaceae (the cabbage family), and Euphorbiaceae (the spurge family) – also use C4.
There are very few trees which use C4. Only a handful are known: Paulownia, seven Hawaiian Euphorbia species and a few desert shrubs that reach the size and shape of trees with age.[23][24]
Converting C3 plants to C4
Given the advantages of C4, a group of scientists from institutions around the world are working on the C4 Rice Project to produce a strain of rice, naturally a C3 plant, that uses the C4 pathway by studying the C4 plants maize and Brachypodium.[25] As rice is the world's most important human food—it is the staple food for more than half the planet—having rice that is more efficient at converting sunlight into grain could have significant global benefits towards improving food security. The team claim C4 rice could produce up to 50% more grain—and be able to do it with less water and nutrients.[26][27][28]
The researchers have already identified genes needed for C4 photosynthesis in rice and are now looking towards developing a prototype C4 rice plant. In 2012, the Government of the United Kingdom along with the Bill & Melinda Gates Foundation provided US$14 million over three years towards the C4 Rice Project at the International Rice Research Institute.[29]
References
- Slack, C. R.; Hatch, M. D. (1967). "Comparative studies on the activity of carboxylases and other enzymes in relation to the new pathway of photosynthetic carbon dioxide fixation in tropical grasses". The Biochemical Journal. 103 (3): 660–665. doi:10.1042/bj1030660. PMC 1270465. PMID 4292834.
- Nickell, Louis G. (1993). "A tribute to Hugo P. Kortschak: The man, the scientist and the discoverer of C4 photosynthesis". Photosynthesis Research. 35 (2): 201–204. doi:10.1007/BF00014751. PMID 24318687.
- Hatch, Marshall D. (2002). "C(4) photosynthesis: Discovery and resolution". Photosynthesis Research. 73 (1–3): 251–6. doi:10.1023/A:1020471718805. PMID 16245128.
- Laetsch (1971). Hatch; Osmond; Slatyer (eds.). Photosynthesis and Photorespiration. New York, Wiley-Interscience.
- Freitag, H.; Stichler, W. (2000). "A remarkable new leaf type with unusual photosynthetic tissue in a central Asiatic genus of Chenopodiaceae". Plant Biology. 2 (2): 154–160. doi:10.1055/s-2000-9462.
- Voznesenskaya, Elena; Vincent R. Franceschi; Olavi Kiirats; Elena G. Artyusheva; Helmut Freitag; Gerald E. Edwards (2002). "Proof of C4 photosynthesis without Kranz anatomy in Bienertia cycloptera (Chenopodiaceae)". The Plant Journal. 31 (5): 649–662. doi:10.1046/j.1365-313X.2002.01385.x. PMID 12207654. S2CID 14742876.
- Akhani, Hossein; Barroca, João; Koteeva, Nuria; Voznesenskaya, Elena; Franceschi, Vincent; Edwards, Gerald; Ghaffari, Seyed Mahmood; Ziegler, Hubert (2005). "Bienertia sinuspersici (Chenopodiaceae): A New Species from Southwest Asia and Discovery of a Third Terrestrial C4 Plant Without Kranz Anatomy". Systematic Botany. 30 (2): 290–301. doi:10.1600/0363644054223684.
- Akhani, H.; Chatrenoor, T.; Dehghani, M.; Khoshravesh, R.; Mahdavi, P.; Matinzadeh, Z. (2012). "A new species of Bienertia (Chenopodiaceae) from Iranian salt deserts: a third species of the genus and discovery of a fourth terrestrial C4 plant without Kranz anatomy". Plant Biosystems. 146: 550–559. doi:10.1080/11263504.2012.662921.
- Holaday, A. S.; Bowes, G. (1980). "C4 Acid Metabolism and Dark CO
2 Fixation in a Submersed Aquatic Macrophyte (Hydrilla verticillata)". Plant Physiology. 65 (2): 331–335. doi:10.1104/pp.65.2.331. PMC 440321. PMID 16661184. - Sage, Rowan; Monson, Russell (1999). "7". C4 Plant Biology. pp. 228–229. ISBN 978-0-12-614440-6.
- Sage, Rowan F. (2004-02-01). "The evolution of C4 photosynthesis". New Phytologist. 161 (2): 341–370. doi:10.1111/j.1469-8137.2004.00974.x. ISSN 1469-8137.
- Williams, B. P.; Johnston, I. G.; Covshoff, S.; Hibberd, J. M. (September 2013). "Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis". eLife. 2: e00961. doi:10.7554/eLife.00961. PMC 3786385. PMID 24082995.
- Osborne, C. P.; Beerling, D. J. (2006). "Nature's green revolution: the remarkable evolutionary rise of C4 plants". Philosophical Transactions of the Royal Society B: Biological Sciences. 361 (1465): 173–194. doi:10.1098/rstb.2005.1737. PMC 1626541. PMID 16553316.
- Edwards, E. J.; Smith, S. A. (2010). "Phylogenetic analyses reveal the shady history of C4 grasses". Proceedings of the National Academy of Sciences. 107 (6): 2532–2537. Bibcode:2010PNAS..107.2532E. doi:10.1073/pnas.0909672107. PMC 2823882. PMID 20142480.
- Osborne, C. P.; Freckleton, R. P. (2009). "Ecological selection pressures for C4 photosynthesis in the grasses". Proceedings of the Royal Society B: Biological Sciences. 276 (1663): 1753–1760. doi:10.1098/rspb.2008.1762. PMC 2674487. PMID 19324795.
- Bond, W. J.; Woodward, F. I.; Midgley, G. F. (2005). "The global distribution of ecosystems in a world without fire". New Phytologist. 165 (2): 525–538. doi:10.1111/j.1469-8137.2004.01252.x. PMID 15720663.
- Kellogg, Elizabeth A. (2013). "C4 photosynthesis". Current Biology. 23 (14): R594–R599. doi:10.1016/j.cub.2013.04.066. PMID 23885869.
- Sage, Rowan F. (2016-07-01). "A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: species number, evolutionary lineages, and Hall of Fame". Journal of Experimental Botany. 67 (14): 4039–4056. doi:10.1093/jxb/erw156. ISSN 0022-0957. PMID 27053721.
- Grass Phylogeny Working Group II (2012). "New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins". New Phytologist. 193 (2): 304–312. doi:10.1111/j.1469-8137.2011.03972.x. hdl:2262/73271. PMID 22115274.

- Sage, Rowan; Monson, Russell (1999). "16". C4 Plant Biology. pp. 551–580. ISBN 978-0-12-614440-6.
- Zhu, X.-G.; Long, S.-P.; Ort, D. R. (2008). "What is the maximum efficiency with which photosynthesis can convert solar energy into biomass?". Current Opinion in Biotechnology. 19 (2): 153–159. doi:10.1016/j.copbio.2008.02.004. PMID 18374559.
- Kadereit, G.; Borsch, T.; Weising, K.; Freitag, H. (2003). "Phylogeny of Amaranthaceae and Chenopodiaceae and the Evolution of C4 Photosynthesis". International Journal of Plant Sciences. 164 (6): 959–986. doi:10.1086/378649.
- Icka, Pirro; Damo, Robert; Icka, Engjëllushe (2016-10-27). "Paulownia tomentosa, a fast growing timber". Annals "Valahia" University of Targoviste - Agriculture. 10 (1): 14–19. doi:10.1515/agr-2016-0003.
- Sage, RF; Sultmanis, S (2016). "Why are there no C4 forests?". J Plant Physiol. 203: 55–68. doi:10.1016/j.jplph.2016.06.009. PMID 27481816.
- Slewinski (2012). "Scarecrow Plays a Role in Establishing Kranz Anatomy in Maize Leaves". Plant & Cell Physiology. 53 (12): 2030–7. doi:10.1093/pcp/pcs147. PMID 23128603.
- Gilles van Kote (2012-01-24). "Researchers aim to flick the high-carbon switch on rice". The Guardian. Retrieved 2012-11-10.
- Von Caemmerer, S.; Quick, W. P.; Furbank, R. T. (2012). "The Development of C4 Rice: Current Progress and Future Challenges". Science. 336 (6089): 1671–1672. Bibcode:2012Sci...336.1671V. doi:10.1126/science.1220177. PMID 22745421.
- Hibberd, J. M.; Sheehy, J. E.; Langdale, J. A. (2008). "Using C4 photosynthesis to increase the yield of rice—rationale and feasibility". Current Opinion in Plant Biology. 11 (2): 228–231. doi:10.1016/j.pbi.2007.11.002. PMID 18203653.
- Hasan, Munawan (2012-11-06). "C4 rice project gets financial boost". The News. Retrieved 2012-11-10.