170
89
Credits to Calvin's Hobbies for nudging my challenge idea in the right direction.
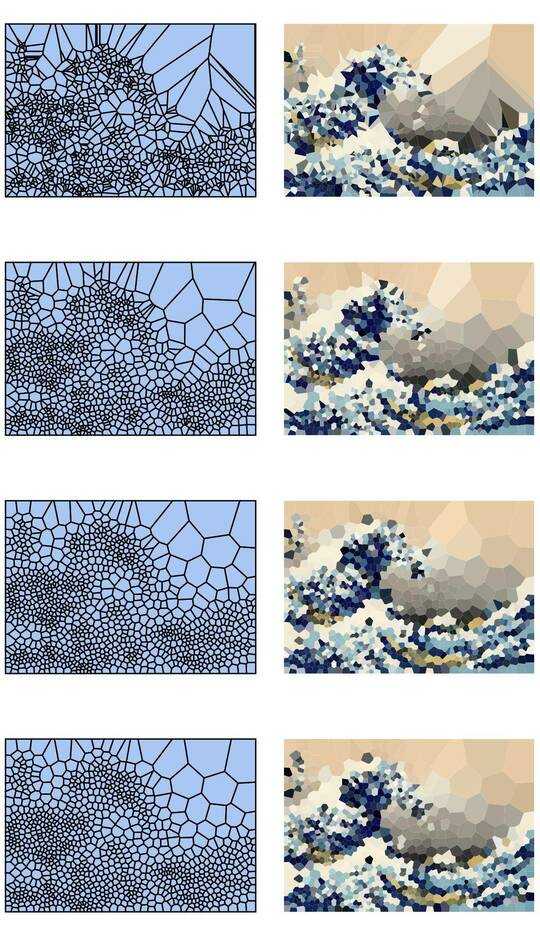
Consider a set of points in the plane, which we will call sites, and associate a colour with each site. Now you can paint the entire plane by colouring each point with the colour of the closest site. This is called a Voronoi map (or Voronoi diagram). In principle, Voronoi maps can be defined for any distance metric, but we'll simply use the usual Euclidean distance r = √(x² + y²). (Note: You do not necessarily have to know how to compute and render one of these to compete in this challenge.)
Here is an example with 100 sites:
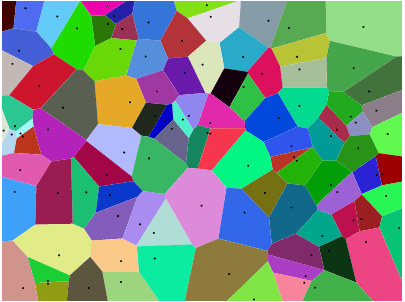
If you look at any cell, then all points within that cell are closer to the corresponding site than to any other site.
Your task is to approximate a given image with such a Voronoi map. You're given the image in any convenient raster graphics format, as well as an integer N. You should then produce up to N sites, and a colour for each site, such that the Voronoi map based on these sites resembles the input image as closely as possible.
You can use the Stack Snippet at the bottom of this challenge to render a Voronoi map from your output, or you can render it yourself if you prefer.
You may use built-in or third-party functions to compute a Voronoi map from a set of sites (if you need to).
This is a popularity contest, so the answer with the most net votes wins. Voters are encouraged to judge answers by
- how well the original images and their colours are approximated.
- how well the algorithm works on different kinds of images.
- how well the algorithm works for small N.
- whether the algorithm adaptively clusters points in regions of the image that require more detail.
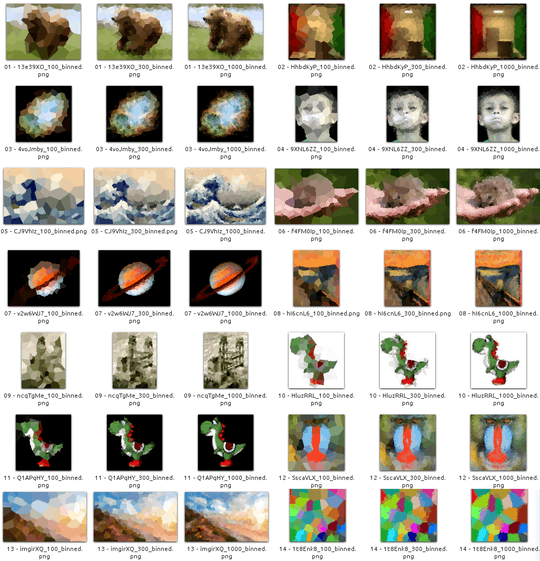
Test Images
Here are a few images to test your algorithm on (some of our usual suspects, some new ones). Click the pictures for larger versions.
The beach in the first row was drawn by Olivia Bell, and included with her permission.
If you want an extra challenge, try Yoshi with a white background and get his belly line right.
You can find all of these test images in this imgur gallery where you can download all of them as a zip file. The album also contains a random Voronoi diagram as another test. For reference, here is the data that generated it.
Please include example diagrams for a variety of different images and N, e.g. 100, 300, 1000, 3000 (as well as pastebins to some of the corresponding cell specifications). You may use or omit black edges between the cells as you see fit (this may look better on some images than on others). Do not include the sites though (except in a separate example maybe if you want to explain how your site placement works, of course).
If you want to show a large number of results, you can create a gallery over on imgur.com, to keep the size of the answers reasonable. Alternatively, put thumbnails in your post and make them links to larger images, like I did in my reference answer. You can get the small thumbnails by appending s to the file name in the imgur.com link (e.g. I3XrT.png -> I3XrTs.png). Also, feel free to use other test images, if you find something nice.
Renderer
Paste your output into the following Stack Snippet to render your results. The exact list format is irrelevant, as long as each cell is specified by 5 floating point numbers in the order x y r g b, where x and y are the coordinates of the cell's site, and r g b are the red, green and blue colour channels in the range 0 ≤ r, g, b ≤ 1.
The snippet provides options to specify a line width of the cell edges, and whether or not the cell sites should be shown (the latter mostly for debugging purposes). But note that the output is only re-rendered when the cell specifications change - so if you modify some of the other options, add a space to the cells or something.
function draw() {
document.getElementById("output").innerHTML = svg
}
function drawMap() {
var cells = document.getElementById("cells").value;
var showSites = document.getElementById("showSites").checked;
var showCells = document.getElementById("showCells").checked;
var lineWidth = parseFloat(document.getElementById("linewidth").value);
var width = parseInt(document.getElementById("width").value);
var height = parseInt(document.getElementById("height").value);
var e = prefix.replace('{{WIDTH}}', width)
.replace('{{HEIGHT}}', height);
cells = cells.match(/(?:\d*\.\d+|\d+\.\d*|\d+)(?:e-?\d+)?/ig);
var sitesDom = '';
var sites = []
for (i = 0; i < cells.length; i+=5)
{
x = parseFloat(cells[i]);
y = parseFloat(cells[i+1]);
r = Math.floor(parseFloat(cells[i+2])*255);
g = Math.floor(parseFloat(cells[i+3])*255);
b = Math.floor(parseFloat(cells[i+4])*255);
sitesDom += '<circle cx="' + x + '" + cy="' + y + '" r="1" fill="black"/>';
sites.push({ x: x, y: y, r: r, g: g, b: b });
}
if (showCells)
{
var bbox = { xl: 0, xr: width, yt: 0, yb: height };
var voronoi = new Voronoi();
var diagram = voronoi.compute(sites, bbox);
for (i = 0; i < diagram.cells.length; ++i)
{
var cell = diagram.cells[i];
var site = cell.site;
var coords = '';
for (var j = 0; j < cell.halfedges.length; ++j)
{
var vertex = cell.halfedges[j].getStartpoint();
coords += ' ' + vertex.x + ',' + vertex.y;
}
var color = 'rgb('+site.r+','+site.g+','+site.b+')';
e += '<polygon fill="'+color+'" points="' + coords + '" stroke="' + (lineWidth ? 'black' : color) + '" stroke-width="'+Math.max(0.6,lineWidth)+'"/>';
}
}
if (showSites)
e += sitesDom;
e += suffix;
e += '<br> Using <b>'+sites.length+'</b> cells.';
svg = e;
draw();
}
var prefix = '<svg width="{{WIDTH}}" height="{{HEIGHT}}">',
suffix = "</svg>",
scale = 0.95,
offset = 225,
radius = scale*offset,
svg = "";svg {
position: relative;
}<script src="http://www.raymondhill.net/voronoi/rhill-voronoi-core.js"></script>
Line width: <input id="linewidth" type="text" size="1" value="0" />
<br>
Show sites: <input id="showSites" type="checkbox" />
<br>
Show cells: <input id="showCells" type="checkbox" checked="checked" />
<br>
<br>
Dimensions: <input id="width" type="text" size="1" value="400" /> x <input id="height" type="text" size="1" value="300" />
<br>
Paste cell specifications here
<br>
<textarea id="cells" cols="60" rows="10" oninput='drawMap()'></textarea>
<br>
<br>
<div id="output"></div>Massive credits to Raymond Hill for writing this really nice JS Voronoi library.


































































































































































































































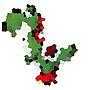











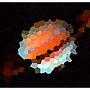













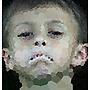


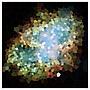




































































How do we verify we have done it the best way? – frogeyedpeas – 2015-05-16T18:51:36.513
5
@frogeyedpeas By looking at the votes you get. ;) This is a popularity contest. There is not necessarily the best way to do. The idea is that you try to do it as well as you can, and the votes will reflect whether people agree that you've done a good job. There is a certain amount of subjectivity in these, admittedly. Have a look at the related challenges I linked, or at this one. You'll see that there is usually a wide variety of approaches but the voting system helps the better solutions bubble up to the top and decide on a winner.
– Martin Ender – 2015-05-16T18:55:08.1203Olivia approves of the approximations of her beach submitted thus far. – Alex A. – 2015-05-17T21:41:14.490
3@AlexA. Devon approves of some of the approximations of his face submitted thus far. He's not a big fan of any of the n=100 versions ;) – Geobits – 2015-05-18T18:58:16.843
1@Geobits: He'll understand when he's older. – Alex A. – 2015-05-18T20:32:40.597
1Here's a page about a centroidal voronoi-based stippling technique. A good source of inspiration (the related master thesis has a nice discussion of possible improvements to the algorithm). – Job – 2015-05-19T11:54:04.663
Did I make a comment about the use of the term "distance measure" and how it should be "metric" or am I going mental? I can't find the comment! – Alec Teal – 2015-05-19T19:06:21.013
@AlecTeal You did, and I fixed it, and I replied... and I saw you were online since my reply, so I assumed you'd read it and deleted both comments as they were obsolete. ;) – Martin Ender – 2015-05-19T19:08:01.780
I had linked to the definition because.... I have a maths background, but it might be the first time someone has seen a formal notion of "distance" and their "metrics" - that's why I included the link. You're using just 3 properties to generate this map. I am also quite interested in seeing what the pictures look like with different metrics.... http://www.maths.kisogo.com/index.php?title=Metric_space is the link BTW
– Alec Teal – 2015-05-19T19:24:38.867Is this just a puzzle or do you also think this could be an approach for a new type of compressed images? To me it looks like storing the coordinates and colors of the sites would need much less information than storing the image pixel by pixel. – Thomas Weller – 2015-05-21T21:00:30.897
@ThomasWeller I just thought it's a fun challenge that is both challenging and would yield varied and interesting results. But the same thing has been discussed when I posted it in Hacker News, so maybe you can weigh in there.
– Martin Ender – 2015-05-21T21:03:09.447