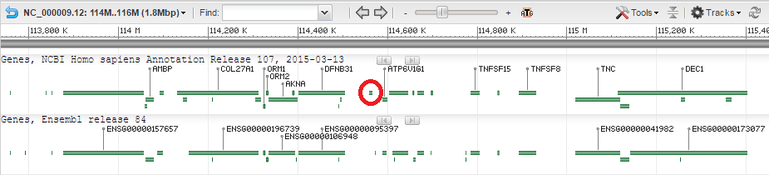
Transmembrane protein 268
Transmembrane protein 268 is a protein that in humans is encoded by TMEM268 gene. The protein is a transmembrane protein of 342 amino acids long with eight alternative splice variants. The protein has been identified in organisms from the common fruit fly to primates. To date, there has been no protein expression found in organisms simpler than insects.
| C9orf91 | |
|---|---|
| Identifiers | |
| Symbol | C9orf91 |
| RefSeq | NP_694590.2 |
| UniProt | Q5VZI3 |
| Other data | |
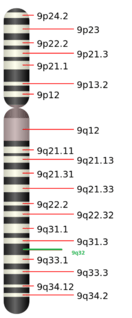
| Locus | Chr. 9 q32 |
Gene
mRNA
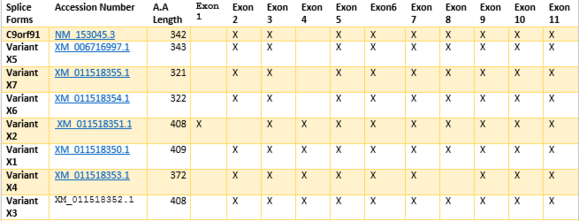
Eight different alternative splice variants for TMEM268 have been found.[3] The exon Sequence for the combinations shown indicate combined regions. Amino acid length differs due to variants in nucleotide additions or deletions within exon regions, conferring different splice variants with similar sequence combinations.
Protein
General Characteristics
TMEM268 has seven variants. NP_694590.2 is the variant that is the most studied. An ExPasy result indicates TMEM268 has an isoelectric point at 5.19 and a molecular weight around 37.6 kdal.[4] TMEM268 is a member of a domain of unknown function, DUF4481.[5] The region is located within 37 and 328 in the amino acid sequence. BLAST results indicate there are no paralogs within humans, zebra fish, and fruit flies.
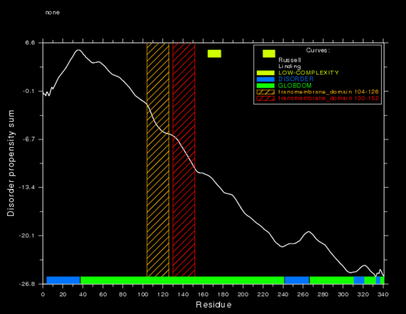
Transmembrane Characteristics
.png)
Two predicted transmembrane regions are on the polypeptide, located at amino acids 104-125 and 130-152 respectively.[7] The SAPS tool on the San Diego Super Computing Biology Workbench garnered protein structural characteristics.[8]
An hypothesis for transmembrane direction is the N terminus and C terminus to remain within the cell, and the loop to stick out of the membrane into the cytosol, presented in the adjacent image.
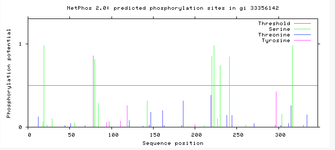
Evidence for the transmembrane direction stems from phosphorylation site predictions. Net Phos 2.0 results for TMEM268 indicate regions of phosphorylation that are present in areas around the transmembrane region. This indicates the N terminus and C terminus are going to be facing into the cell, providing support to the transmembrane direction.
Protein Expression
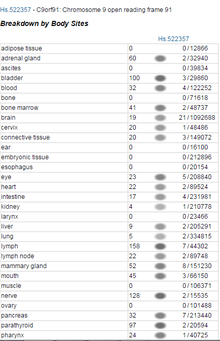
The protein was found to be expressed in numerous different tissues. The ETS Profile for TMEM268 indicates the gene is expressed ubiquitously within the body.[10] In support, there has been experimental evidence using antibody staining to show various tissue types that express TMEM268.[11]
A recent study linked TMEM268 as a lysosome transporter.[12] The exact function within the lysosome and interactions with other molecules, however, is not known.
Associated Disease
Due to limited characterization, linkage to associated diseases is not well known. Techniques to view SNPs have been used in numerous diseases to determine loci influence. In spondylocostal dysostosis and spondyloarthritis, both were found to have SNPs that had indicated TMEM268 as a possible factor to the diseases[13][14]
Interacting Proteins
Interacting proteins were found with c-REL(via affinity chromotography and yeast-two hybrid) and ELAV1, via a yeast-two hybrid.[15][16] REL is associated to be a proto-oncogene that is from a family of transcription factors. It is associated with B-cell proliferation. ELAV1 is associated with binding to mRNA in order to help increase stability of transcripts.
Homology
Ortholog identity and similarity
Identity of orthologous domains within TMEM268 in other species with a 40% identity and above is shown below. The DUFF4481 start shown on the image retains conservation throughout time. Of interest, areas of transmembrane region are not as conserved as thought, indicating that specific amino acids may not be a vital component towards the protein's function.
Conservation within species
TMEM268 is conserved in many organisms. Orthologs have been found in mammals, birds, reptiles, amphibians, sharks, fish, and a few insects. It was not found within plants, bacteria, or archea. Amino acid length tends to increase within orthologs as divergence from humans goes up based on BLAST search results for similar proteins.
References
- "I-TASSER server for protein structure and function prediction". zhanglab.ccmb.med.umich.edu. Retrieved 2016-05-08.
- mieg@ncbi.nlm.nih.gov, Danielle Thierry-Mieg and Jean Thierry-Mieg, NCBI/NLM/NIH. "AceView: Gene:C9orf91, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView". www.ncbi.nlm.nih.gov. Retrieved 2016-04-28.
- "NCBI Gene".
- "ExPASy isoelectric point".
- "transmembrane protein C9orf91 [Homo sapiens] - Protein - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2016-04-28.
- "GlobPlot 2.3 - Predictor of intrinsic protein disorder & globularity". globplot.embl.de. Retrieved 2016-04-28.
- "C9orf91 - Transmembrane protein C9orf91 - Homo sapiens (Human) - C9orf91 gene & protein". www.uniprot.org. Retrieved 2016-04-28.
- "SDSC Biology Workbench". workbench.sdsc.edu. Retrieved 2016-04-28.
- "GlobPlot 2.3 - Predictor of intrinsic protein disorder & globularity". globplot.embl.de. Retrieved 2016-04-28.
- "EST Profile - Hs.522357". www.ncbi.nlm.nih.gov. Retrieved 2016-04-28.
- "Tissue expression of C9orf91 - Summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2016-04-28.
- Chapel A, Kieffer-Jaquinod S, Sagné C, Verdon Q, Ivaldi C, Mellal M, et al. (June 2013). "An extended proteome map of the lysosomal membrane reveals novel potential transporters". Molecular & Cellular Proteomics. 12 (6): 1572–88. doi:10.1074/mcp.M112.021980. PMC 3675815. PMID 23436907.
- Zinovieva E, Kadi A, Letourneur F, Cagnard N, Izac B, Vigier A, et al. (July 2011). "Systematic candidate gene investigations in the SPA2 locus (9q32) show an association between TNFSF8 and susceptibility to spondylarthritis". Arthritis and Rheumatism. 63 (7): 1853–9. doi:10.1002/art.30377. PMID 21480186.
- Sparrow DB, McInerney-Leo A, Gucev ZS, Gardiner B, Marshall M, Leo PJ, et al. (April 2013). "Autosomal dominant spondylocostal dysostosis is caused by mutation in TBX6". Human Molecular Genetics. 22 (8): 1625–31. doi:10.1093/hmg/ddt012. PMID 23335591.
- Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, Kuwano Y, et al. (May 2009). "Ubiquitin-mediated proteolysis of HuR by heat shock". The EMBO Journal. 28 (9): 1271–82. doi:10.1038/emboj.2009.67. PMC 2683047. PMID 19322201.
- Wang J, Huo K, Ma L, Tang L, Li D, Huang X, et al. (October 2011). "Toward an understanding of the protein interaction network of the human liver". Molecular Systems Biology. 7: 536. doi:10.1038/msb.2011.67. PMC 3261708. PMID 21988832.