Talairach coordinates
Talairach coordinates, also known as Talairach space, is a 3-dimensional coordinate system (known as an 'atlas') of the human brain, which is used to map the location of brain structures independent from individual differences in the size and overall shape of the brain. It is still common to use Talairach coordinates in functional brain imaging studies and to target transcranial stimulation of brain regions.[1] However, alternative methods such as the MNI Coordinate System (originated at the Montreal Neurological Institute and Hospital) have largely replaced Talairach for stereotaxy and other procedures.[2]
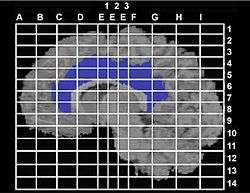
History
The coordinate system was first created by neurosurgeons Jean Talairach and Gabor Szikla in their work on the Talairach Atlas in 1967, creating a standardized grid for neurosurgery.[3] The grid was based on the idea that distances to lesions in the brain are proportional to overall brain size (i.e., the distance between two structures is larger in a larger brain). In 1988 a second edition of the Talairach Atlas came out that was coauthored by Tournoux, and it is sometimes known as the Talairach-Tournoux system. This atlas was based on single post-mortem dissection of a human brain.[4]
The Talairach Atlas uses Brodmann areas as the labels for brain regions.[5]
Description
The Talairach coordinate system is defined by making two anchors, the anterior commissure and posterior commissure, lie on a straight horizontal line.[6] Since these two points lie on the midsagittal plane, the coordinate system is completely defined by requiring this plane to be vertical. Distances in Talairach coordinates are measured from the anterior commissure as the origin (as defined in the 1998 edition). The y-axis points posterior and anterior to the commissures, the left and right is the x-axis, and the z-axis is in the ventral-dorsal (down and up) directions.[7] Once the brain is reoriented to these axes, the researchers must also outline the six cortical outlines of the brain: anterior, posterior, left, right, inferior, and superior.[8] In the 1967 atlas the left is with positive coordinates while in the 1988 atlas the left has negative coordinates.
By defining standard anatomical landmarks that could be identified on different subjects (the anterior and posterior commissures), it became easier to spatially warp an individual brain image obtained through Magnetic Resonance Imaging (MRI), positron emission tomography (PET) and other imaging methods to this standard Talairach space. One can then make inferences about tissue identity at a specific location by referring to the atlas.
Brodmann regions in neuroscience
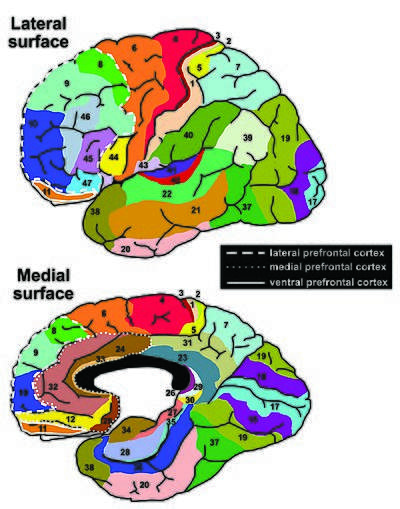
The Brodmann area is an illustration of a cytoarchitectonic map of the human brain that was published by Korbinan Brodmann in his 1909 monogram. Brodmann's map splits the cerebral cortex into 43 differing parts, which become visible in cell-body stained histological sections. Years later, a large group of neuroscientists still utilize Brodmann's map for the localization of neuroimaging data that is obtained in living human brains.[9]
Brodmann in relation to Talairach coordinates
Some neuroimaging techniques, in fact, purported the use of Brodmann's area as a guideline for Talairach coordinates. Further, these technologies showcase that experimental tasks in a common reference space become possible through imaging the living human brain by registering function and having architectonic data performed and defined.
Brodmann's map proved useful in varying neuroimaging software packages and stereotaxic atlases, such as the Talairach atlas. This atlas also serves as a demonstration of the inherent problems (i.e., impressions of matches between areal borders and sulcal landmarks may lead to wrong conclusions in terms of localization of cytoarchitectonic borders or the usage of Brodmann's map without knowledge of the text that accompanies the drawing misleading researchers to false conclusions).[9]
Conversion to other coordinate systems
Montreal Neurological Institute (MNI) templates
Another common atlas for the human brain is the Montreal Neurological Institute and Hospital (MNI) coordinate system, which is the template used for SPM and the International Consortium for Brain Mapping. Most neuroimaging software packages are able to convert from Talairach to MNI coordinates.
However, disparities between MNI and Talairach coordinates can impede the comparison of results across different studies. This problem is most prevalent in situations where coordinate disparities should be corrected to reduce error, such as coordinate-based meta-analyses. There is a possibility that these disparities can be assuaged through the Lancaster transform, which can be adopted in order to minimize the variability in the literature regarding spatial normalization strategies.[10]
Non-linear registration
Non-linear registration is the process of digitalizing Talairach coordinates and generating a non-linear map in an attempt to compensate for actual shape differences between the two, which would result in more accurate coordinate transformations.
The Talairach atlas is still commonly used in terms of the neuroimaging techniques that are available, but the lack of a three-dimensional model of the original brain makes it difficult for researchers to map locations from three-dimensional anatomical MRI images to the atlas automatically. Previous methods such as MNI have tried to assuage this issue through linear and piecewise mapping between the Talairach and the MNI template, but can only account for differences in overall brain orientation and size and thus cannot correctly account for actual shape differences.[11]
Optimized high-resolution brain template (HRBT)
Current target brains are not suitable for current research (i.e., are average, can only be used in low-resolution MRI target brain mapping studies or are single brain). Optimized high-resolution brain template (HRBT), a high-resolution MRI target brain, is a technique that can aid in the issues named latter. This optimization can be performed to help reduce individual anatomical biases of the original ICBM HBRT. The optimized HRBT is more adept at anatomically matching groups of brains.[12]
References
- Handbook of Medical Imaging: Processing and Analysis Management. Academic Press. 9 October 2000. p. 565. ISBN 978-0-08-053310-0.
- Russell A. Poldrack; Jeanette A. Mumford; Thomas E. Nichols (22 August 2011). Handbook of Functional MRI Data Analysis. Cambridge University Press. p. 17. ISBN 978-1-139-49836-4.
- Talairach, Jean; Szikla, G. (1967). "Atlas of stereotactic concepts to the surgery of epilepsy". Cite journal requires
|journal=(help) - Lazar, Nicole (10 June 2008). The Statistical Analysis of Functional MRI Data. Springer. pp. 88–. ISBN 978-0-387-78191-4.
- Brent Vogt (4 June 2009). Cingulate Neurobiology and Disease. Oxford University Press. p. 4. ISBN 978-0-19-856696-0.
- Guang-Zhong Yang; Tianzi Jiang (11 August 2004). Medical Imaging and Augmented Reality: Second International Workshop, MIAR 2004, Beijing, China, August 19-20, 2004, Proceedings. Springer. pp. 179. ISBN 978-3-540-22877-6.
- Klaus D. Toennies (4 February 2012). Guide to Medical Image Analysis: Methods and Algorithms. Springer. p. 326. ISBN 978-1-4471-2751-2.
- Bruce L. Miller; Jeffrey L. Cummings (2007). The Human Frontal Lobes: Functions and Disorders. Guilford Press. p. 173. ISBN 978-1-59385-329-7.
- Zilles, Karl; Amunts, Katrin (2010-01-04). "Centenary of Brodmann's map — conception and fate". Nature Reviews Neuroscience. 11 (2): 139–145. doi:10.1038/nrn2776. ISSN 1471-003X.
- Laird, Angela R.; Robinson, Jennifer L.; McMillan, Kathryn M.; Tordesillas-Gutiérrez, Diana; Moran, Sarah T.; Gonzales, Sabina M.; Ray, Kimberly L.; Franklin, Crystal; Glahn, David C.; Fox, Peter T.; Lancaster, Jack L. (June 2010). "Comparison of the disparity between Talairach and MNI coordinates in functional neuroimaging data: Validation of the Lancaster transform". NeuroImage. 51 (2): 677–683. doi:10.1016/j.neuroimage.2010.02.048. PMC 2856713. PMID 20197097.
- Lacadie, Cheryl M.; Fulbright, Robert K.; Rajeevan, Nallakkandi; Constable, R. Todd; Papademetris, Xenophon (August 2008). "More accurate Talairach coordinates for neuroimaging using non-linear registration". NeuroImage. 42 (2): 717–725. doi:10.1016/j.neuroimage.2008.04.240. PMC 2603575.
- Kochunov, P.; Lancaster, J.; Thompson, P.; Toga, A.W.; Brewer, P.; Hardies, J.; Fox, P. (October 2002). "An Optimized Individual Target Brain in the Talairach Coordinate System". NeuroImage. 17 (2): 922–927. doi:10.1006/nimg.2002.1084.