Supersecondary structure
A supersecondary structure is a compact three-dimensional protein structure of several adjacent elements of a secondary structure that is smaller than a protein domain or a subunit. Supersecondary structures can act as nucleations in the process of protein folding.
Examples
Helix supersecondary structures

Helix hairpin
A helix hairpin, also known as an alpha-alpha hairpin, is composed of two antiparallel alpha helices connected by a loop of two or more residues. True to its name, it resembles a hairpin. A longer loop has a greater number of possible conformations. If short strands connect the helices, then the individual helices will pack together through their hydrophobic residues. The function of a helix hairpin is unknown; however, a four helix bundle is composed of two helix hairpins, which have important ligand binding sites.[1]
Helix corner
A helix corner, also called an alpha-alpha corner, has two alpha helices almost at right angles to each other connected by a short 'loop'. This loop is formed from a hydrophobic residue. The function of a helix corner is unknown.[1]
Helix-loop-helix
The helix-loop-helix structure has two helices connected by a 'loop'. These are fairly common and usually bind ligands. For example, calcium binds with the carboxyl groups of the side chains within the loop region between the helices.[1]
Helix-turn-helix
The helix-turn-helix motif is important for DNA binding and is therefore in many DNA binding proteins.[1]
Beta sheet supersecondary structures

Beta hairpin
A beta hairpin is a common supersecondary motif composed of two anti-parallel beta strands connected by a loop. The structure resembles a hairpin and is often found in globular proteins.
The loop between the beta strands can range anywhere from 2 to 16 residues. However, most loops contain less than seven residues.[2] Residues in beta hairpins with loops of 2, 3, or 4 residues have distinct conformations. However, a wide range of conformations can be seen in longer loops, which are sometimes referred to as 'random coils'. A beta-meander consists of consecutive antiparallel-beta strands linked by hairpins.[3]
Two residue loops are called beta turns or reverse turns. Type I' and Type II' reverse turns occur most frequently because they have less steric hindrance than Type I and Type II turns. The function of beta hairpins is unknown.[2]
Beta corner
A beta hairpin has two antiparallel beta strands that are at about a 90 degree angle to each other. It is formed by a beta hairpin changing direction with one strand having a glycine residue and the other strand having a beta bulge. Beta corners have no known function.[2]
Greek key motif
A Greek key motif has four features:
- Four sequentially connected beta strands are adjacent to, much not necessarily geometrically aligned with, each other.
- The beta sheet is anti-parallel, and alternate strands run in opposite directions.
- The first strand and last strand are next to each other and bonded by hydrogen bonds.
- Connecting loops can be long and include other secondary structures.
The Greek key motif has its name because the structure looks like the pattern seen on Greek urns. This motif has no known function.
Other
β-sheets (composed of multiple hydrogen-bonded individual β-strands) are sometimes considered a secondary or supersecondary structure.
Mixed supersecondary structures
Beta-alpha-beta motifs
A beta-alpha-beta motif is composed of two beta strands joined by an alpha helix through connecting loops. The beta strands are parallel, and the helix is also almost parallel to the strands. This structure can be seen in almost all proteins with parallel strands. The loops connecting the beta strands and alpha helix can vary in length and often binds ligands.
Beta-alpha-beta helices can be either left-handed or right-handed. When viewed from the N-terminal side of the beta strandes, so that one strand is on top of the other, a left-handed beta-alpha-beta motif has the alpha helix on the left side of the beta strands. The more common right-handed motif would have an alpha helix on the right side of the plane containing the beta strands.[4]
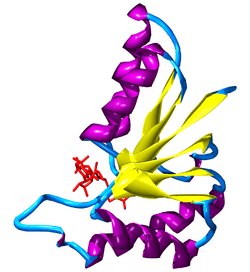
Rossman fold
Rossman folds, named after Michael Rossman, consist of 3 beta strands and 2 helices in an alternating fashion: beta strand, helix, beta strand, helix, beta strand. This motif tends to reverse the direction of the chain within a protein. Rossman folds have an important biological function in binding nucleotides such as NAD within most dehydrogenases.[4]
References
- "Helix Supersecondary Structures". biomedapps.curtin.edu.au. Retrieved 2020-01-31.
- "Beta sheet supersecondary structures". biomedapps.curtin.edu.au. Retrieved 2020-01-31.
- Cooper, J. (1995-01-26). "Supersecondary Structure". VSNS-PPS Course Material. Retrieved 2020-03-14.
- "Mixed Beta Strand and Helix Structures". biomedapps.curtin.edu.au. Retrieved 2020-01-31.
Further reading
- Chiang YS, Gelfand TI, Kister AE, Gelfand IM (2007). "New classification of supersecondary structures of sandwich-like proteins uncovers strict patterns of strand assemblage". Proteins. 68 (4): 915–921. doi:10.1002/prot.21473. PMID 17557333.