Sulfoglycolysis
Sulfoglycolysis is a catabolic process in primary metabolism in which sulfoquinovose (6-deoxy-6-sulfonato-glucose) is metabolized to produce energy and carbon-building blocks.[1] Sulfoglycolysis pathways occur in a wide variety of organisms, and enable key steps in the degradation of sulfoquinovosyl diacylglycerol (SQDG), a sulfolipid found in plants and cyanobacteria into sulfite and sulfate. Sulfoglycolysis converts sulfoquinovose (C6H12O8S−) into pyruvate CH3COCOO− + H+. The free energy is used to form the high-energy molecules ATP (adenosine triphosphate) and NADH (reduced nicotinamide adenine dinucleotide). Unlike glycolysis, all known sulfoglycolysis pathways convert only half the carbon content of sulfoquinovose into pyruvate; the remained is excreted as a C3-sulfonate: 2,3-dihydroxypropanesulfonate (DHPS) or sulfolactate (SL).
Two sulfoglycolytic processes are known. The sulfoglycolytic Embden-Meyerhof-Parnas (sulfo-EMP) pathway, first identified in Escherichia coli, involves the degradation of sulfoquinovose to 2,3-dihydroxypropanesulfonate (DHPS),[2] and shares similarity with the Embden-Meyerhof-Parnas glycolysis pathway. The sulfoglycolytic Entner-Doudoroff (sulfo-ED) pathway, first identified in Pseudomonas putida SQ1, involves the degradation of sulfoquinovose to sulfolactate,[3] and shares similarity to the Entner-Doudoroff pathway of glycolysis. Both pathways lead to the production of dihydroxyacetone phosphate in processes that consume ATP. Energy is formed in later stages through the 'pay-off' phase of glycolysis through substrate-level phosphorylation to produce ATP and NADH.
Growth of bacteria on sulfoquinovose and its glycosides
A range of bacteria can grow on sulfoquinovose or its glycosides as sole carbon source. E. coli can grow on sulfoquinovose,[2] methyl α-sulfoquinovoside and α-sulfoquinovosyl glycerol.[4] Growth on sulfoquinovosyl glycerol is both faster and leads to higher cell density than for growth on sulfoquinovose.[4] Pseudomonas aeruginosa strain SQ1,[5] Klebsiella sp. strain ABR11,[6] Klebsiella oxytoca TauN1,[5] and Agrobacterium sp. strain ABR2[6] can grow on sulfoquinovose as sole carbon source. A strain of Flavobacterium was identified that could grow on methyl α-sulfoquinovoside.[7]
Production of sulfoquinovose and its mutarotation
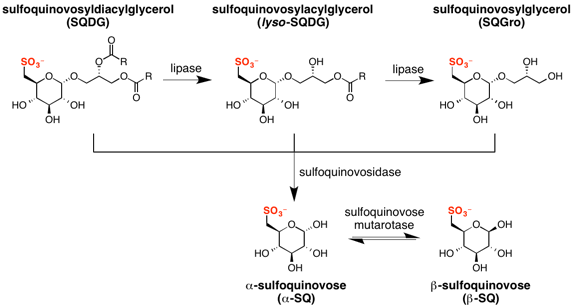
Sulfoquinovose is rarely found in its free form in nature; rather it occurs predominantly as a glycoside, SQDG. SQDG can be deacylated to form lyso-SQDG and sulfoquinovosylglycerol (SQGro).[8][9][10] Sulfoquinovose is obtained from SQ glycosides by the action of sulfoquinovosidases, which are glycoside hydrolases that can hydrolyse the glycosidic linkage in SQDG, or its deacylated form, sulfoquinovosyl glycerol (SQGro).[11] The first sulfoquinovosidase identified was YihQ from Escherichia coli. It exhibits a preference for the naturally occurring 2’R-SQGro.[4] Sulfoquinovosidases cleave SQ glycosides with retention of configuration, initially forming α-sulfoquinovose. Sulfoglycolysis encoding operons contain gene sequences encoding aldose-1-epimerases that act as sulfoquinovose mutarotases, catalyzing the interconversion of the α and β anomers of sulfoquinovose.[12]
Sulfo-EMP pathway
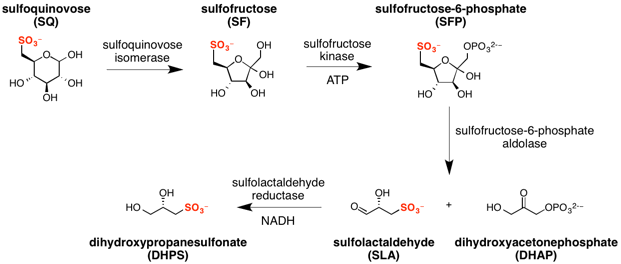
The major steps in the sulfo-EMP pathway[2] are:
- isomerization of sulfoquinovose to sulfofructose (catalyzed by sulfoquinovose isomerase);
- phosphorylation of sulfofructose to sulfofructose-1-phosphate (catalyzed by sulfofructose kinase and using ATP as a co-factor);
- retro-aldol cleavage of sulfofructose-1-phosphate to afford dihydroxyacetone phosphate and (S)-sulfolactaldehyde (catalyzed by sulfofructose-1-phosphate aldolase);
- reduction of sulfolactaldehyde to (S)-2,3-dihydroxypropane-1-sulfonate (catalyzed by sulfolactaldehyde reductase and using NADH as a co-factor).
Expression of proteins within the sulfo-EMP operon of E. coli is regulated by a transcription factor termed CsqR (formerly YihW).[13] CsqR binds to DNA sites within the operon encoding the sulfo-EMP pathway, functioning as a repressor. SQ and SQGro (but not lactose, glucose or galactose) function as derepressors of CsqR.
Sulfo-ED pathway
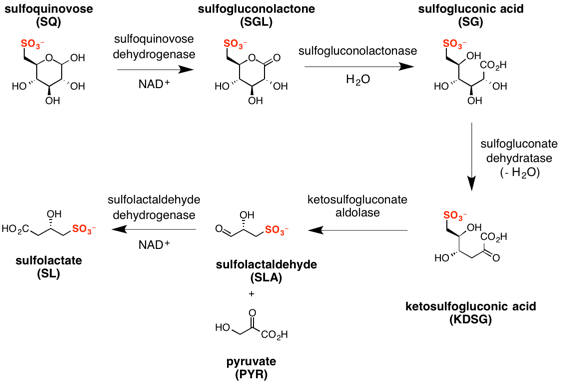
The major steps in the sulfo-ED pathway[3] are:
- oxidation of sulfoquinovose to sulfogluconolactone (catalyzed by sulfoquinovose dehydrogenase with NAD+ co-factor);
- hydrolysis of sulfogluconolactone to sulfogluconate acid (catalyzed by sulfogluconolactonase with water);
- dehydration of sulfogluconic acid to 2-keto-3,6-dideoxy-6-sulfogluconate (catalyzed by sulfogluconate dehydratase);
- retro-aldol cleavage of 2-keto-3,6-dideoxy-6-sulfogluconate to give pyruvate and (S)-sulfolactaldehyde (catalyzed by sulfoketogluconate dehydrogenase with NAD+ co-factor);
- oxidation of sulfolactaldehyde to (S)-sulfolactate (catalyzed by sulfolactaldehyde dehydrogenase with NAD+ co-factor).
Degradation of DHPS and SL
The C3 sulfonates DHPS and SL are metabolized for their carbon content, as well as to mineralize their sulfur content.[1] Metabolism of DHPS typically involves conversion to SL. Metabolism of SL can occur in several ways including:
- elimination of sulfite to afford pyruvate;
- oxidation to sulfopyruvate, transamination to cysteate, and elimination of sulfite to afford pyruvate and ammonia;
- oxidation to sulfopyruvate, decarboxylation to sulfoacetaldehyde, and phosphorylation to afford acetylphosphate and sulfite.
See also
References
- Goddard-Borger ED, Williams SJ (February 2017). "Sulfoquinovose in the biosphere: occurrence, metabolism and functions". The Biochemical Journal. 474 (5): 827–849. doi:10.1042/BCJ20160508. PMID 28219973.
- Denger K, Weiss M, Felux AK, Schneider A, Mayer C, Spiteller D, Huhn T, Cook AM, Schleheck D (March 2014). "Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle". Nature. 507 (7490): 114–7. Bibcode:2014Natur.507..114D. doi:10.1038/nature12947. PMID 24463506.
- Felux AK, Spiteller D, Klebensberger J, Schleheck D (August 2015). "Entner-Doudoroff pathway for sulfoquinovose degradation in Pseudomonas putida SQ1". Proceedings of the National Academy of Sciences of the United States of America. 112 (31): E4298–305. Bibcode:2015PNAS..112E4298F. doi:10.1073/pnas.1507049112. PMC 4534283. PMID 26195800.
- Abayakoon, Palika; Jin, Yi; Lingford, James P.; Petricevic, Marija; John, Alan; Ryan, Eileen; Wai-Ying Mui, Janice; Pires, Douglas E.V.; Ascher, David B. (2018-09-05). "Structural and Biochemical Insights into the Function and Evolution of Sulfoquinovosidases". ACS Central Science. 4 (9): 1266–1273. doi:10.1021/acscentsci.8b00453. ISSN 2374-7943. PMC 6161063. PMID 30276262.
- Denger K, Huhn T, Hollemeyer K, Schleheck D, Cook AM (March 2012). "Sulfoquinovose degraded by pure cultures of bacteria with release of C3-organosulfonates: complete degradation in two-member communities". FEMS Microbiology Letters. 328 (1): 39–45. doi:10.1111/j.1574-6968.2011.02477.x. PMID 22150877.
- Roy AB, Hewlins MJ, Ellis AJ, Harwood JL, White GF (November 2003). "Glycolytic breakdown of sulfoquinovose in bacteria: a missing link in the sulfur cycle". Applied and Environmental Microbiology. 69 (11): 6434–41. doi:10.1128/AEM.69.11.6434-6441.2003. PMC 262304. PMID 14602597.
- Martelli HL, Benson AA (October 1964). "Sulfocarbohydrate metabolism. I. bacterial production and utilization of sulfoacetate". Biochimica et Biophysica Acta. 93: 169–71. doi:10.1016/0304-4165(64)90272-7. PMID 14249144.
- Wolfersberger MG, Pieringer RA (January 1974). "Metabolism of sulfoquinovosyl diglyceride in Chlorella pyrenoidosa by sulfoquinovosyl monoglyceride: fatty acyl CoA acyltransferase and sulfoquinovosyl glyceride: fatty acyl ester hydrolase pathways". Journal of Lipid Research. 15 (1): 1–10. PMID 4359538.
- Gupta SD, Sastry PS (December 1987). "Metabolism of the plant sulfolipid--sulfoquinovosyldiacylglycerol: degradation in animal tissues". Archives of Biochemistry and Biophysics. 259 (2): 510–9. doi:10.1016/0003-9861(87)90517-0. PMID 3426241.
- Andersson L, Bratt C, Arnoldsson KC, Herslöf B, Olsson NU, Sternby B, Nilsson A (June 1995). "Hydrolysis of galactolipids by human pancreatic lipolytic enzymes and duodenal contents". Journal of Lipid Research. 36 (6): 1392–400. PMID 7666015.
- Speciale G, Jin Y, Davies GJ, Williams SJ, Goddard-Borger ED (April 2016). "YihQ is a sulfoquinovosidase that cleaves sulfoquinovosyl diacylglyceride sulfolipids" (PDF). Nature Chemical Biology. 12 (4): 215–7. doi:10.1038/nchembio.2023. PMID 26878550.
- Abayakoon P, Lingford JP, Jin Y, Bengt C, Davies GJ, Yao S, Goddard-Borger ED, Williams SJ (April 2018). "Discovery and characterization of a sulfoquinovose mutarotase using kinetic analysis at equilibrium by exchange spectroscopy". The Biochemical Journal. 475 (7): 1371–1383. doi:10.1042/BCJ20170947. PMC 5902678. PMID 29535276.
- Shimada, Tomohiro; Yamamoto, Kaneyoshi; Nakano, Masahiro; Watanabe, Hiroki; Schleheck, David; Ishihama, Akira (29 October 2018). "Regulatory role of CsqR (YihW) in transcription of the genes for catabolism of the anionic sugar sulfoquinovose (SQ) in Escherichia coli K-12". Microbiology. 165 (1): 78–89. doi:10.1099/mic.0.000740. PMID 30372406.