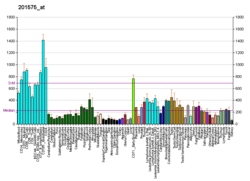
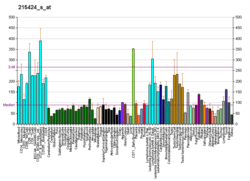
SNW1
SNW domain-containing protein 1 is a protein that in humans is encoded by the SNW1 gene.[5][6][7]
Function
This gene, a member of the SNW gene family, encodes a coactivator that enhances transcription from some Pol II promoters. This coactivator can bind to the ligand-binding domain of the vitamin D receptor and to retinoid receptors to enhance vitamin D-, retinoic acid-, estrogen-, and glucocorticoid-mediated gene expression. It can also function as a splicing factor by interacting with poly(A)-binding protein 2 to directly control the expression of muscle-specific genes at the transcriptional level. Finally, the protein may be involved in oncogenesis since it interacts with a region of SKI oncoproteins that is required for transforming activity.[7]
Interactions
SNW1 has been shown to interact with:
- CIR,[8]
- Calcitriol receptor,[6][9]
- Histone deacetylase 2,[8]
- Mothers against decapentaplegic homolog 2,[10]
- Mothers against decapentaplegic homolog 3,[10]
- NOTCH1[11][12]
- Nuclear receptor co-repressor 2,[8][11]
- Nuclear receptor coactivator 1,[9]
- PABPN1,[13]
- RBPJ,[8][11]
- Retinoblastoma protein,[14] and
- SKI protein.[15][16][10]
References
- GRCh38: Ensembl release 89: ENSG00000100603 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000021039 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Folk P, Půta F, Krpejsová L, Blahůsková A, Markos A, Rabino M, Dottin RP (Nov 1996). "The homolog of chromatin binding protein Bx42 identified in Dictyostelium". Gene. 181 (1–2): 229–31. doi:10.1016/S0378-1119(96)00483-0. PMID 8973337.
- Baudino TA, Kraichely DM, Jefcoat SC, Winchester SK, Partridge NC, MacDonald PN (Jun 1998). "Isolation and characterization of a novel coactivator protein, NCoA-62, involved in vitamin D-mediated transcription". The Journal of Biological Chemistry. 273 (26): 16434–41. doi:10.1074/jbc.273.26.16434. PMID 9632709.
- "Entrez Gene: SNW1 SNW domain containing 1".
- Zhou S, Fujimuro M, Hsieh JJ, Chen L, Hayward SD (Feb 2000). "A role for SKIP in EBNA2 activation of CBF1-repressed promoters". Journal of Virology. 74 (4): 1939–47. doi:10.1128/jvi.74.4.1939-1947.2000. PMC 111672. PMID 10644367.
- Zhang C, Baudino TA, Dowd DR, Tokumaru H, Wang W, MacDonald PN (Nov 2001). "Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription". The Journal of Biological Chemistry. 276 (44): 40614–20. doi:10.1074/jbc.M106263200. PMID 11514567.
- Leong GM, Subramaniam N, Figueroa J, Flanagan JL, Hayman MJ, Eisman JA, Kouzmenko AP (May 2001). "Ski-interacting protein interacts with Smad proteins to augment transforming growth factor-beta-dependent transcription". The Journal of Biological Chemistry. 276 (21): 18243–8. doi:10.1074/jbc.M010815200. PMID 11278756.
- Zhou S, Fujimuro M, Hsieh JJ, Chen L, Miyamoto A, Weinmaster G, Hayward SD (Apr 2000). "SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC To facilitate NotchIC function". Molecular and Cellular Biology. 20 (7): 2400–10. doi:10.1128/mcb.20.7.2400-2410.2000. PMC 85419. PMID 10713164.
- Beatus P, Lundkvist J, Oberg C, Pedersen K, Lendahl U (Jun 2001). "The origin of the ankyrin repeat region in Notch intracellular domains is critical for regulation of HES promoter activity". Mechanisms of Development. 104 (1–2): 3–20. doi:10.1016/s0925-4773(01)00373-2. PMID 11404076.
- Kim YJ, Noguchi S, Hayashi YK, Tsukahara T, Shimizu T, Arahata K (May 2001). "The product of an oculopharyngeal muscular dystrophy gene, poly(A)-binding protein 2, interacts with SKIP and stimulates muscle-specific gene expression". Human Molecular Genetics. 10 (11): 1129–39. doi:10.1093/hmg/10.11.1129. PMID 11371506.
- Prathapam T, Kühne C, Banks L (Dec 2002). "Skip interacts with the retinoblastoma tumor suppressor and inhibits its transcriptional repression activity". Nucleic Acids Research. 30 (23): 5261–8. doi:10.1093/nar/gkf658. PMC 137971. PMID 12466551.
- Prathapam T, Kühne C, Hayman M, Banks L (Sep 2001). "Ski interacts with the evolutionarily conserved SNW domain of Skip". Nucleic Acids Research. 29 (17): 3469–76. doi:10.1093/nar/29.17.3469. PMC 55893. PMID 11522815.
- Dahl R, Wani B, Hayman MJ (Mar 1998). "The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42". Oncogene. 16 (12): 1579–86. doi:10.1038/sj.onc.1201687. PMID 9569025.
Further reading
- Dahl R, Wani B, Hayman MJ (Mar 1998). "The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42". Oncogene. 16 (12): 1579–86. doi:10.1038/sj.onc.1201687. PMID 9569025.
- Zhou S, Fujimuro M, Hsieh JJ, Chen L, Hayward SD (Feb 2000). "A role for SKIP in EBNA2 activation of CBF1-repressed promoters". Journal of Virology. 74 (4): 1939–47. doi:10.1128/JVI.74.4.1939-1947.2000. PMC 111672. PMID 10644367.
- Zhou S, Fujimuro M, Hsieh JJ, Chen L, Miyamoto A, Weinmaster G, Hayward SD (Apr 2000). "SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC To facilitate NotchIC function". Molecular and Cellular Biology. 20 (7): 2400–10. doi:10.1128/MCB.20.7.2400-2410.2000. PMC 85419. PMID 10713164.
- Leong GM, Subramaniam N, Figueroa J, Flanagan JL, Hayman MJ, Eisman JA, Kouzmenko AP (May 2001). "Ski-interacting protein interacts with Smad proteins to augment transforming growth factor-beta-dependent transcription". The Journal of Biological Chemistry. 276 (21): 18243–8. doi:10.1074/jbc.M010815200. PMID 11278756.
- Kim YJ, Noguchi S, Hayashi YK, Tsukahara T, Shimizu T, Arahata K (May 2001). "The product of an oculopharyngeal muscular dystrophy gene, poly(A)-binding protein 2, interacts with SKIP and stimulates muscle-specific gene expression". Human Molecular Genetics. 10 (11): 1129–39. doi:10.1093/hmg/10.11.1129. PMID 11371506.
- Beatus P, Lundkvist J, Oberg C, Pedersen K, Lendahl U (Jun 2001). "The origin of the ankyrin repeat region in Notch intracellular domains is critical for regulation of HES promoter activity". Mechanisms of Development. 104 (1–2): 3–20. doi:10.1016/S0925-4773(01)00373-2. PMID 11404076.
- Zhou S, Hayward SD (Sep 2001). "Nuclear localization of CBF1 is regulated by interactions with the SMRT corepressor complex". Molecular and Cellular Biology. 21 (18): 6222–32. doi:10.1128/MCB.21.18.6222-6232.2001. PMC 87339. PMID 11509665.
- Zhang C, Baudino TA, Dowd DR, Tokumaru H, Wang W, MacDonald PN (Nov 2001). "Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription". The Journal of Biological Chemistry. 276 (44): 40614–20. doi:10.1074/jbc.M106263200. PMID 11514567.
- Prathapam T, Kühne C, Hayman M, Banks L (Sep 2001). "Ski interacts with the evolutionarily conserved SNW domain of Skip". Nucleic Acids Research. 29 (17): 3469–76. doi:10.1093/nar/29.17.3469. PMC 55893. PMID 11522815.
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ (Apr 2002). "Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis". RNA. 8 (4): 426–39. doi:10.1017/S1355838202021088. PMC 1370266. PMID 11991638.
- Wardell SE, Boonyaratanakornkit V, Adelman JS, Aronheim A, Edwards DP (Aug 2002). "Jun dimerization protein 2 functions as a progesterone receptor N-terminal domain coactivator". Molecular and Cellular Biology. 22 (15): 5451–66. doi:10.1128/MCB.22.15.5451-5466.2002. PMC 133955. PMID 12101239.
- Prathapam T, Kühne C, Banks L (Dec 2002). "Skip interacts with the retinoblastoma tumor suppressor and inhibits its transcriptional repression activity". Nucleic Acids Research. 30 (23): 5261–8. doi:10.1093/nar/gkf658. PMC 137971. PMID 12466551.
- Petersen HH, Hilpert J, Militz D, Zandler V, Jacobsen C, Roebroek AJ, Willnow TE (Feb 2003). "Functional interaction of megalin with the megalinbinding protein (MegBP), a novel tetratrico peptide repeat-containing adaptor molecule". Journal of Cell Science. 116 (Pt 3): 453–61. doi:10.1242/jcs.00243. PMID 12508107.
- Barry JB, Leong GM, Church WB, Issa LL, Eisman JA, Gardiner EM (Mar 2003). "Interactions of SKIP/NCoA-62, TFIIB, and retinoid X receptor with vitamin D receptor helix H10 residues". The Journal of Biological Chemistry. 278 (10): 8224–8. doi:10.1074/jbc.C200712200. PMID 12529369.
- Zhang C, Dowd DR, Staal A, Gu C, Lian JB, van Wijnen AJ, Stein GS, MacDonald PN (Sep 2003). "Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing". The Journal of Biological Chemistry. 278 (37): 35325–36. doi:10.1074/jbc.M305191200. PMID 12840015.
- Leong GM, Subramaniam N, Issa LL, Barry JB, Kino T, Driggers PH, Hayman MJ, Eisman JA, Gardiner EM (Mar 2004). "Ski-interacting protein, a bifunctional nuclear receptor coregulator that interacts with N-CoR/SMRT and p300". Biochemical and Biophysical Research Communications. 315 (4): 1070–6. doi:10.1016/j.bbrc.2004.02.004. PMID 14985122.