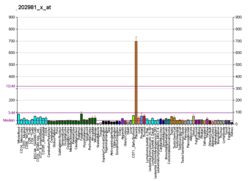
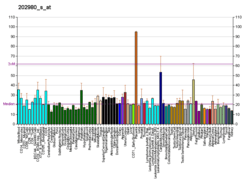
SIAH1
E3 ubiquitin-protein ligase SIAH1 is an enzyme that in humans is encoded by the SIAH1 gene.[5][6]
Function
This gene encodes for a polypeptide structure that is a member of the seven in absentia homolog (SIAH) family. The protein is an E3 ligase and is involved in ubiquitination and proteasome-mediated degradation of specific proteins. The activity of this ubiquitin ligase has been implicated in the development of certain forms of Parkinson's disease, the regulation of the cellular response to hypoxia and induction of apoptosis. Alternative splicing results in several additional transcript variants, some encoding different isoforms and others that have not been fully characterized.[7]
Interactions
SIAH1 has been shown to interact with:
gollark: And that pagination will be hobbled for some inexplicable reason to 5 pages.
gollark: Don't be ridiculous. The TJ09 works in the cover of darkness.
gollark: What are they worth *now* then?
gollark: Also, what are purple siyats actually worth these days? I got 4.
gollark: So, encouragement for people to uselessly log on a bit?
References
- GRCh38: Ensembl release 89: ENSG00000196470 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000036840 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Hu G, Chung YL, Glover T, Valentine V, Look AT, Fearon ER (November 1997). "Characterization of human homologs of the Drosophila seven in absentia (sina) gene". Genomics. 46 (1): 103–11. doi:10.1006/geno.1997.4997. PMID 9403064.
- Hu G, Zhang S, Vidal M, Baer JL, Xu T, Fearon ER (October 1997). "Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway". Genes & Development. 11 (20): 2701–14. doi:10.1101/gad.11.20.2701. PMC 316613. PMID 9334332.
- "Entrez Gene: SIAH1 seven in absentia homolog 1 (Drosophila)".
- Liu J, Stevens J, Rote CA, Yost HJ, Hu Y, Neufeld KL, White RL, Matsunami N (May 2001). "Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein". Molecular Cell. 7 (5): 927–36. doi:10.1016/S1097-2765(01)00241-6. PMID 11389840.
- Matsuzawa S, Takayama S, Froesch BA, Zapata JM, Reed JC (May 1998). "p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1". The EMBO Journal. 17 (10): 2736–47. doi:10.1093/emboj/17.10.2736. PMC 1170614. PMID 9582267.
- Matsuzawa SI, Reed JC (May 2001). "Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses". Molecular Cell. 7 (5): 915–26. doi:10.1016/S1097-2765(01)00242-8. PMID 11389839.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Venables JP, Dalgliesh C, Paronetto MP, Skitt L, Thornton JK, Saunders PT, Sette C, Jones KT, Elliott DJ (July 2004). "SIAH1 targets the alternative splicing factor T-STAR for degradation by the proteasome". Human Molecular Genetics. 13 (14): 1525–34. doi:10.1093/hmg/ddh165. PMID 15163637.
- Germani A, Bruzzoni-Giovanelli H, Fellous A, Gisselbrecht S, Varin-Blank N, Calvo F (December 2000). "SIAH-1 interacts with alpha-tubulin and degrades the kinesin Kid by the proteasome pathway during mitosis". Oncogene. 19 (52): 5997–6006. doi:10.1038/sj.onc.1204002. PMID 11146551.
- Susini L, Passer BJ, Amzallag-Elbaz N, Juven-Gershon T, Prieur S, Privat N, Tuynder M, Gendron MC, Israël A, Amson R, Oren M, Telerman A (December 2001). "Siah-1 binds and regulates the function of Numb". Proceedings of the National Academy of Sciences of the United States of America. 98 (26): 15067–72. doi:10.1073/pnas.261571998. PMC 64984. PMID 11752454.
- Okabe H, Satoh S, Furukawa Y, Kato T, Hasegawa S, Nakajima Y, Yamaoka Y, Nakamura Y (June 2003). "Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1". Cancer Research. 63 (12): 3043–8. PMID 12810624.
- Relaix F, Wei Xj, Li W, Pan J, Lin Y, Bowtell DD, Sassoon DA, Wu X (February 2000). "Pw1/Peg3 is a potential cell death mediator and cooperates with Siah1a in p53-mediated apoptosis". Proceedings of the National Academy of Sciences of the United States of America. 97 (5): 2105–10. doi:10.1073/pnas.040378897. PMC 15761. PMID 10681424.
- Tiedt R, Bartholdy BA, Matthias G, Newell JW, Matthias P (August 2001). "The RING finger protein Siah-1 regulates the level of the transcriptional coactivator OBF-1". The EMBO Journal. 20 (15): 4143–52. doi:10.1093/emboj/20.15.4143. PMC 149178. PMID 11483517.
- Germani A, Prabel A, Mourah S, Podgorniak MP, Di Carlo A, Ehrlich R, Gisselbrecht S, Varin-Blank N, Calvo F, Bruzzoni-Giovanelli H (December 2003). "SIAH-1 interacts with CtIP and promotes its degradation by the proteasome pathway". Oncogene. 22 (55): 8845–51. doi:10.1038/sj.onc.1206994. PMID 14654780.
- Zhou Y, Li L, Liu Q, Xing G, Kuai X, Sun J, Yin X, Wang J, Zhang L, He F (May 2008). "E3 ubiquitin ligase SIAH1 mediates ubiquitination and degradation of TRB3". Cellular Signalling. 20 (5): 942–8. doi:10.1016/j.cellsig.2008.01.010. PMID 18276110.
Further reading
- Maruyama K, Sugano S (January 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Nemani M, Linares-Cruz G, Bruzzoni-Giovanelli H, Roperch JP, Tuynder M, Bougueleret L, Cherif D, Medhioub M, Pasturaud P, Alvaro V, der Sarkissan H, Cazes L, Le Paslier D, Le Gall I, Israeli D, Dausset J, Sigaux F, Chumakov I, Oren M, Calvo F, Amson RB, Cohen D, Telerman A (August 1996). "Activation of the human homologue of the Drosophila sina gene in apoptosis and tumor suppression". Proceedings of the National Academy of Sciences of the United States of America. 93 (17): 9039–42. doi:10.1073/pnas.93.17.9039. PMC 38591. PMID 8799150.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (October 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Block KL, Vornlocher HP, Hershey JW (November 1998). "Characterization of cDNAs encoding the p44 and p35 subunits of human translation initiation factor eIF3". The Journal of Biological Chemistry. 273 (48): 31901–8. doi:10.1074/jbc.273.48.31901. PMID 9822659.
- Hu G, Fearon ER (January 1999). "Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins". Molecular and Cellular Biology. 19 (1): 724–32. doi:10.1128/mcb.19.1.724. PMC 83929. PMID 9858595.
- Tanikawa J, Ichikawa-Iwata E, Kanei-Ishii C, Nakai A, Matsuzawa S, Reed JC, Ishii S (May 2000). "p53 suppresses the c-Myb-induced activation of heat shock transcription factor 3". The Journal of Biological Chemistry. 275 (20): 15578–85. doi:10.1074/jbc.M000372200. PMID 10747903.
- Medhioub M, Vaury C, Hamelin R, Thomas G (September 2000). "Lack of somatic mutation in the coding sequence of SIAH1 in tumors hemizygous for this candidate tumor suppressor gene". International Journal of Cancer. 87 (6): 794–7. doi:10.1002/1097-0215(20000915)87:6<794::AID-IJC5>3.0.CO;2-B. PMID 10956387.
- Matsuzawa SI, Reed JC (May 2001). "Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses". Molecular Cell. 7 (5): 915–26. doi:10.1016/S1097-2765(01)00242-8. PMID 11389839.
- Boehm J, He Y, Greiner A, Staudt L, Wirth T (August 2001). "Regulation of BOB.1/OBF.1 stability by SIAH". The EMBO Journal. 20 (15): 4153–62. doi:10.1093/emboj/20.15.4153. PMC 149152. PMID 11483518.
- Wheeler TC, Chin LS, Li Y, Roudabush FL, Li L (March 2002). "Regulation of synaptophysin degradation by mammalian homologues of seven in absentia". The Journal of Biological Chemistry. 277 (12): 10273–82. doi:10.1074/jbc.M107857200. PMID 11786535.
- Maeda A, Yoshida T, Kusuzaki K, Sakai T (February 2002). "The characterization of the human Siah-1 promoter(1)". FEBS Letters. 512 (1–3): 223–6. doi:10.1016/S0014-5793(02)02265-2. PMID 11852084.
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N (March 2002). "An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22". Genomics. 79 (3): 285–96. doi:10.1006/geno.2002.6718. PMID 11863358.
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.