SHANK2
SH3 and multiple ankyrin repeat domains protein 2 is a protein that in humans is encoded by the SHANK2 gene.[5][6] Two alternative splice variants, encoding distinct isoforms, are reported. Additional splice variants exist but their full-length nature has not been determined.[6]
Function
This gene encodes a protein that is a member of the Shank family of synaptic proteins that may function as molecular scaffolds in the postsynaptic density (PSD). Shank proteins contain multiple domains for protein-protein interaction, including ankyrin repeats, an SH3 domain, a PSD-95/Dlg/ZO-1 domain, a sterile alpha motif domain, and a proline-rich region. This particular family member contains a PDZ domain, a consensus sequence for cortactin SH3 domain-binding peptides and a sterile alpha motif. The alternative splicing demonstrated in Shank genes has been suggested as a mechanism for regulating the molecular structure of Shank and the spectrum of Shank-interacting proteins in the PSDs of adult and developing brain.[6]
It is thought that SHANK2 might play a role in synaptogenesis by attaching metabotropic glutamate receptors (mGluRs) to an existing pool of NMDA receptors (NMDA-R), bylinking to the NMDA-R through PSD-95, and the mGluRs through HOMER1.[7] An alternative hypothesis is that the Homer/Shank/GKAP/PSD-95 assembly mediates physical association of the NMDAR with IP3R/RYR and intracellular Ca2+ stores.
Interactions
SHANK2 has been shown to interact with:
Associations with neuropsychiatric disease
Mutations in SHANK2 have been associated with autism spectrum disorder (ASD) and schizophrenia. In particular, heterozygous loss-of-function mutations have a near-complete penetrance in ASD.[13] Neurons generated from people with ASD and SHANK2 mutations develop larger dendritic trees and more synaptic connections than those from healthy controls.[14]
References
- GRCh38: Ensembl release 89: ENSG00000162105 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000037541 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, Kim E (November 1999). "Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development". J Biol Chem. 274 (41): 29510–8. doi:10.1074/jbc.274.41.29510. PMID 10506216.
- "Entrez Gene: SHANK2 SH3 and multiple ankyrin repeat domains 2".
- Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED (June 2002). "ProSAP/Shank proteins - a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease". J. Neurochem. 81 (5): 903–10. doi:10.1046/j.1471-4159.2002.00931.x. PMID 12065602.
- Park E, Na M, Choi J, Kim S, Lee JR, Yoon J, Park D, Sheng M, Kim E (May 2003). "The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42". J. Biol. Chem. 278 (21): 19220–9. doi:10.1074/jbc.M301052200. PMID 12626503.
- Du Y, Weed SA, Xiong WC, Marshall TD, Parsons JT (October 1998). "Identification of a novel cortactin SH3 domain-binding protein and its localization to growth cones of cultured neurons". Mol. Cell. Biol. 18 (10): 5838–51. PMC 109170. PMID 9742101.
- Naisbitt S, Valtschanoff J, Allison DW, Sala C, Kim E, Craig AM, Weinberg RJ, Sheng M (June 2000). "Interaction of the postsynaptic density-95/guanylate kinase domain-associated protein complex with a light chain of myosin-V and dynein". J. Neurosci. 20 (12): 4524–34. doi:10.1523/JNEUROSCI.20-12-04524.2000. PMC 6772433. PMID 10844022.
- Boeckers TM, Winter C, Smalla KH, Kreutz MR, Bockmann J, Seidenbecher C, Garner CC, Gundelfinger ED (October 1999). "Proline-rich synapse-associated proteins ProSAP1 and ProSAP2 interact with synaptic proteins of the SAPAP/GKAP family". Biochem. Biophys. Res. Commun. 264 (1): 247–52. doi:10.1006/bbrc.1999.1489. PMID 10527873.
- Okamoto PM, Gamby C, Wells D, Fallon J, Vallee RB (Dec 2001). "Dynamin isoform-specific interaction with the shank/ProSAP scaffolding proteins of the postsynaptic density and actin cytoskeleton". J. Biol. Chem. 276 (51): 48458–65. doi:10.1074/jbc.M104927200. PMC 2715172. PMID 11583995.
- Leblond, Claire S.; Nava, Caroline; Polge, Anne; Gauthier, Julie; Huguet, Guillaume; Lumbroso, Serge; Giuliano, Fabienne; Stordeur, Coline; Depienne, Christel; Mouzat, Kevin; Pinto, Dalila (2014-09-04). "Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: A Gradient of Severity in Cognitive Impairments". PLOS Genetics. 10 (9): e1004580. doi:10.1371/journal.pgen.1004580. ISSN 1553-7404. PMC 4154644. PMID 25188300.
- Zaslavsky, Kirill; Zhang, Wen-Bo; McCready, Fraser P.; Rodrigues, Deivid C.; Deneault, Eric; Loo, Caitlin; Zhao, Melody; Ross, P. Joel; El Hajjar, Joelle; Romm, Asli; Thompson, Tadeo (April 2019). "SHANK2 mutations associated with autism spectrum disorder cause hyperconnectivity of human neurons". Nature Neuroscience. 22 (4): 556–564. doi:10.1038/s41593-019-0365-8. ISSN 1546-1726.
Further reading
- Sheng M, Kim E (2000). "The Shank family of scaffold proteins". J. Cell Sci. 113. ( Pt 11): 1851–6. PMID 10806096.
- Andersson B, Wentland MA, Ricafrente JY, Liu W, Gibbs RA (1996). "A "double adaptor" method for improved shotgun library construction". Anal. Biochem. 236 (1): 107–13. doi:10.1006/abio.1996.0138. PMID 8619474.
- Yu W, Andersson B, Worley KC, Muzny DM, Ding Y, Liu W, Ricafrente JY, Wentland MA, Lennon G, Gibbs RA (1997). "Large-scale concatenation cDNA sequencing". Genome Res. 7 (4): 353–8. doi:10.1101/gr.7.4.353. PMC 139146. PMID 9110174.
- Du Y, Weed SA, Xiong WC, Marshall TD, Parsons JT (1998). "Identification of a novel cortactin SH3 domain-binding protein and its localization to growth cones of cultured neurons". Mol. Cell. Biol. 18 (10): 5838–51. PMC 109170. PMID 9742101.
- Zitzer H, Richter D, Kreienkamp HJ (1999). "Agonist-dependent interaction of the rat somatostatin receptor subtype 2 with cortactin-binding protein 1". J. Biol. Chem. 274 (26): 18153–6. doi:10.1074/jbc.274.26.18153. PMID 10373412.
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF (1999). "Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins". Neuron. 23 (3): 583–92. doi:10.1016/S0896-6273(00)80810-7. PMID 10433269.
- Kikuno R, Nagase T, Ishikawa K, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (1999). "Prediction of the coding sequences of unidentified human genes. XIV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro". DNA Res. 6 (3): 197–205. doi:10.1093/dnares/6.3.197. PMID 10470851.
- Boeckers TM, Winter C, Smalla KH, Kreutz MR, Bockmann J, Seidenbecher C, Garner CC, Gundelfinger ED (1999). "Proline-rich synapse-associated proteins ProSAP1 and ProSAP2 interact with synaptic proteins of the SAPAP/GKAP family". Biochem. Biophys. Res. Commun. 264 (1): 247–52. doi:10.1006/bbrc.1999.1489. PMID 10527873.
- Kreienkamp HJ, Zitzer H, Gundelfinger ED, Richter D, Bockers TM (2000). "The calcium-independent receptor for alpha-latrotoxin from human and rodent brains interacts with members of the ProSAP/SSTRIP/Shank family of multidomain proteins". J. Biol. Chem. 275 (42): 32387–90. doi:10.1074/jbc.C000490200. PMID 10964907.
- Kreienkamp HJ, Zitzer H, Richter D (2001). "Identification of proteins interacting with the rat somatostatin receptor subtype 2". J. Physiol. Paris. 94 (3–4): 193–8. doi:10.1016/S0928-4257(00)00204-7. PMID 11087996.
- Okamoto PM, Gamby C, Wells D, Fallon J, Vallee RB (2002). "Dynamin isoform-specific interaction with the shank/ProSAP scaffolding proteins of the postsynaptic density and actin cytoskeleton". J. Biol. Chem. 276 (51): 48458–65. doi:10.1074/jbc.M104927200. PMC 2715172. PMID 11583995.
- Soltau M, Richter D, Kreienkamp HJ (2003). "The insulin receptor substrate IRSp53 links postsynaptic shank1 to the small G-protein cdc42". Mol. Cell. Neurosci. 21 (4): 575–83. doi:10.1006/mcne.2002.1201. PMID 12504591.
- Park E, Na M, Choi J, Kim S, Lee JR, Yoon J, Park D, Sheng M, Kim E (2003). "The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42". J. Biol. Chem. 278 (21): 19220–9. doi:10.1074/jbc.M301052200. PMID 12626503.
- Han W, Kim KH, Jo MJ, Lee JH, Yang J, Doctor RB, Moe OW, Lee J, Kim E, Lee MG (2006). "Shank2 associates with and regulates Na+/H+ exchanger 3". J. Biol. Chem. 281 (3): 1461–9. doi:10.1074/jbc.M509786200. PMID 16293618.
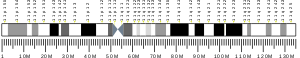
- Taylor TD, Noguchi H, Totoki Y, Toyoda A, Kuroki Y, Dewar K, Lloyd C, Itoh T, Takeda T, Kim DW, She X, Barlow KF, Bloom T, Bruford E, Chang JL, Cuomo CA, Eichler E, FitzGerald MG, Jaffe DB, LaButti K, Nicol R, Park HS, Seaman C, Sougnez C, Yang X, Zimmer AR, Zody MC, Birren BW, Nusbaum C, Fujiyama A, Hattori M, Rogers J, Lander ES, Sakaki Y (2006). "Human chromosome 11 DNA sequence and analysis including novel gene identification". Nature. 440 (7083): 497–500. doi:10.1038/nature04632. PMID 16554811.