Repressilator
The repressilator is a genetic regulatory network consisting of at least one feedback loop with at least three genes, each expressing a protein that represses the next gene in the loop.[1] In biological research, repressilators have been used to build cellular models and understand cell function. There are both artificial and naturally-occurring repressilators. Recently, the naturally-occurring repressilator clock gene circuit in Arabidopsis thaliana (A. thaliana) and mammalian systems have been studied.
Artificial Repressilators
Artificial repressilators were first engineered by Michael Elowitz and Stanislas Leibler in 2000,[2] complementing other research projects studying simple systems of cell components and function. In order to understand and model the design and cellular mechanisms that confers a cell’s function, Elowitz and Leibler created an artificial network consisting of a loop with three transcriptional repressors. This network was designed from scratch to exhibit a stable oscillation that acts like an electrical oscillator system with fixed time periods. The network was implemented in Escherichia coli (E. coli) via recombinant DNA transfer. It was then verified that the engineered colonies did indeed exhibit the desired oscillatory behavior.
The repressilator consists of three genes connected in a feedback loop, such that each gene represses the next gene in the loop and is repressed by the previous gene. In the synthetic insertion into E. Coli, green fluorescent protein (GFP) was used as a reporter so that the behavior of the network could be observed using fluorescence microscopy.
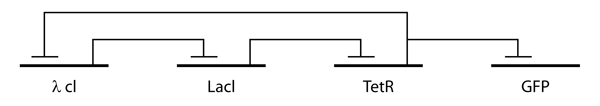
The design of the repressilator was guided by biological and circuit principles with discrete and stochastic models of analysis. Six differential equations were used to model the kinetics of the repressilator system based on protein and mRNA concentrations, as well as appropriate parameter and Hill coefficient values. In the study, Elowitz and Leibler generated figures showing oscillations of repressor proteins, using integration and typical parameter values as well as a stochastic version of the repressilator model using similar parameters. These models were analyzed to determine the values of various rates that would yield a sustained oscillation. It was found that these oscillations were favored by promoters coupled to efficient ribosome binding sites, cooperative transcriptional repressors, and comparable protein and mRNA decay rates.
This analysis motivated two design features which were engineered into the genes. First, promoter regions were replaced with a more efficient hybrid promoter which combined the E. coli phage lambda PL (λ PL) promoter with lac repressor (Lacl) and Tet repressor (TetR) operator sequences. Second, to reduce the disparity between the lifetimes of the repressor proteins and the mRNAs, a carboxy terminal tag based on the ssrA-RNA sequence was added at the 3' end of each repressor gene. This tag is recognized by proteases which target the protein for degradation. The design was implemented using a low-copy plasmid encoding the repressilator and a higher-copy reporter, which were used to transform a culture of E. coli.
Naturally-Occurring Repressilators
Plants
Circadian circuits in plants feature a transcriptional regulatory feedback loop called the repressilator. In the core oscillator loop (outlined in gray) in A. thaliana, light is first sensed by two cryptochromes and five phytochromes. Two transcription factors, Circadian Clock Associated 1 (CCA1) and Late Elongated Hypocotyl (LHY), repress genes associated with evening expression like Timing of CAB expression 1 (TOC1) and activate genes associated with morning expression by binding to their promoters. TOC1, an evening gene, positively regulates CCA1 and LHY via an unknown mechanism.[3] Evening-phased transcription factor CCA1 Hiking Expedition (CHE) and histone demethylase jumonji C domain-containing 5 (JMJD5) directly repress CCA1. Other components have been found to be expressed throughout the day and either directly or indirectly inhibit or activate a consequent element in the circadian circuit, thereby creating a complex, robust and flexible network of feedback loops.[3]
Morning-Phase Expression
The morning-phase expression loop refers to the genes and proteins that regulate rhythms during the day in A. thaliana. The two main genes are LHY and CCA1, which encode LHY and CCA1 transcription factors.[4] These proteins form heterodimers that enter the nucleus and bind to the TOC1 gene promoter, repressing the production of TOC1 protein. When TOC1 protein is expressed, it serves to regulate LHY and CCA1 by inhibition of their transcription. This was later supported in 2012 by Dr. Alexandra Pokhilo, who used computational analyses to show that TOC1 served this role as an inhibitor of LHY and CCA1 expression.[5] The morning loop serves to inhibit hypocotyl elongation, in contrast with the evening-phase loop which promotes hypocotyl elongation. The morning phase loop has shown to be incapable of supporting circadian oscillation when evening-phase expression genes have been mutated,[5] suggesting the interdependency of each component in this naturally-occurring repressilator.
Evening-Phase Expression
Early Flowering 3 (ELF3), Early Flowering 4 (ELF4) and Phytoclock1 (LUX) are the key elements in evening-phased clock gene expression in A. thaliana. They form the evening complex, in which LUX binds to the promoters of Phytochrome Interacting Factor 4 (PIF4) and Phytochrome Interacting Factor 5 (PIF5) and inhibits them.[3] As a result, hypocotyl elongation is repressed in the early-evening. When the inhibition is alleviated late at night, the hypocotyl elongates. Photoperiod flowering is controlled by output gene Gigantea (GI). GI is activated at night and activates the expression of Constans (CO), which activates the expression of Flowering Locus T (FT). FT then causes flowering in long-days.[3]
Mammals
Mammals evolved an endogenous timing mechanism to coordinate both physiology and behavior to the 24 hour period.[6] In 2016, researchers identified a sequence of three subsequent inhibitions within this mechanism that they identified as a repressilator, which is now believed to serve as a major core element of this circadian network. The necessity of this system was established through a series of gene knockouts amongst cryptochrome (Cry), period (Per), and Rev-erb -- core mammalian clock genes whose knockouts lead to arrhythmicity.[6] The model that these researchers generated includes Bmal1 as a driver of E-box mediated transcription, Per2 and Cry1 as early and late E-box repressors, respectively, as well as the D-box regulator Dbp and the nuclear receptor Rev-erb-α. The sequential inhibitions by Rev-erb, Per and Cry1 can generate sustained oscillations, and by clamping all other components except for this repressilator oscillations persisted with similar amplitudes and periods.[6] All oscillating networks seem to involve any combination of these three core genes, as demonstrated in various schematics released by researchers.
Recent Work
The repressilator model has been used to model and study other biological pathways and systems. Since, extensive work into the repressilator’s modeling capacities has been performed. In 2003, the repressilator’s representation and validaton of biological models, being a model with many variables, was performed using the Simpathica system, which verified that the model does indeed oscillate with all of its complexities.
As stated in Elowitz and Leibler’s original work, the ultimate goal for repressilator research is to build an artificial circadian clock that mirrors its natural, endogenous counterpart. This would involve developing an artificial clock with reduced noise and temperature compensation in order to better understand circadian rhythms that can be found in every domain of life.[7] Disruption of circadian rhythms may lead to loss of rhythmicity in metabolic and transcriptional processes, and even quicken the onset of certain neurodegenerative diseases such as Alzheimer's disease.[8] In 2017, oscillators that generated circadian rhythms and were not influenced much by temperature were created in a laboratory.[6]
Pathologically, the repressilator model can be used to model cell growth and abnormalities that may arise, such as those present in cancer cells.[9] In doing so, new treatments may be developed based on circadian activity of cancerous cells. Additionally, in 2016, a research team improved upon the previous design of the repressilator. Following noise (signal processing) analysis, the authors moved the GFP reporter construct onto the repressilator plasmid and removed the ssrA degradation tags from each repressor protein. This extended the period and improved the regularity of the oscillations of the repressilator.[10]
In 2019, a study furthered Elowitz and Leibler's model by improving the repressilator system by achieving a model with a unique steady state and new rate function. This experiment expanded the current knowledge of repression and gene regulation.[11]
Significance
Synthetic Biology
Artificial repressilators were discovered by implanting a synthetic inhibition loop into E. coli. This represented the first implementation of synthetic oscillations into an organism. Further implications of this include the possibility of rescuing mutated components of oscillations synthetically in model organisms.[7]
The artificial repressilator is a milestone of synthetic biology which shows that genetic regulatory networks can be designed and implemented to perform novel functions. However, it was found that the cells' oscillations drifted out of phase after a period of time and the artificial repressilator's activity was influenced by cell growth. The initial experiment[7] therefore gave new appreciation to the circadian clock found in many organisms, as endogenous repressilators are significantly more robust than implanted artificial repressilators. New investigations at the RIKEN Quantitative Biology Center have found that chemical modifications to a single protein molecule could form a temperature independent, self-sustainable oscillator .[12]
Artificial repressilators could potentially aid research and treatments in fields ranging from circadian biology to endocrinology. They are increasingly able to demonstrate the synchronization inherent to natural biological systems and the factors that affect them.[13]
Circadian Biology
A better understanding of the naturally-occurring repressilator in model organisms with endogenous, circadian timings, like A. thaliana, has applications in agriculture, especially in regards to plant rearing and livestock management.[14]
References
- Oliveira, Samuel M. D.; Chandraseelan, Jerome G.; Häkkinen, Antti; Goncalves, Nadia S. M.; Yli-Harja, Olli; Startceva, Sofia; Ribeiro, Andre S. (2015). "Single-cell kinetics of a repressilator when implemented in a single-copy plasmid". Mol. BioSyst. 11 (7): 1939–1945. doi:10.1039/c5mb00012b. PMID 25923804.
- Stanislas Leibler; Elowitz, Michael B. (2000-01-20). "A synthetic oscillatory network of transcriptional regulators". Nature. 403 (6767): 335–338. Bibcode:2000Natur.403..335E. doi:10.1038/35002125. ISSN 1476-4687. PMID 10659856.
- Kay, Steve A.; Nagel, Dawn H. (2012-08-21). "Complexity in the Wiring and Regulation of Plant Circadian Networks". Current Biology. 22 (16): R648–R657. doi:10.1016/j.cub.2012.07.025. ISSN 0960-9822. PMC 3427731. PMID 22917516.
- Hsu, Polly Yingshan; Harmer, Stacey L. (Spring 2014). "Wheels within wheels: the plant circadian system". Trends in Plant Science. 19 (4): 240–249. doi:10.1016/j.tplants.2013.11.007. ISSN 1360-1385. PMC 3976767. PMID 24373845.
- Pokhilko, Alexandra; Fernández, Aurora Piñas; Edwards, Kieron D; Southern, Megan M; Halliday, Karen J; Millar, Andrew J (2012-03-13). "The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops". Molecular Systems Biology. 8: 574. doi:10.1038/msb.2012.6. ISSN 1744-4292. PMC 3321525. PMID 22395476.
- Wu, Lili; Ouyang, Qi; Wang, Hongli (2017-02-02). "Robust network topologies for generating oscillations with temperature-independent periods". PLoS ONE. 12 (2): e0171263. Bibcode:2017PLoSO..1271263W. doi:10.1371/journal.pone.0171263. ISSN 1932-6203. PMC 5289577. PMID 28152061.
- A Synthetic Oscillatory Network of Transcriptional Regulators; Michael Elowitz and Stanislas Leibler; Nature. 2000 Jan 20;403(6767):335-8.
- Homolak, Jan; Mudrovčić, Monika; Vukić, Barbara; Toljan, Karlo (2018-06-21). "Circadian Rhythm and Alzheimer's Disease". Medical Sciences. 6 (3): 52. doi:10.3390/medsci6030052. ISSN 2076-3271. PMC 6164904. PMID 29933646.
- Szymańska, Zuzanna; Cytowski, Maciej; Mitchell, Elaine; Macnamara, Cicely K.; Chaplain, Mark A. J. (May 2018). "Computational Modelling of Cancer Development and Growth: Modelling at Multiple Scales and Multiscale Modelling". Bulletin of Mathematical Biology. 80 (5): 1366–1403. doi:10.1007/s11538-017-0292-3. hdl:10023/14364. ISSN 1522-9602. PMID 28634857.
- Potvin-Trottier, Laurent; Lord, Nathan D.; Vinnicombe, Glenn; Paulsson, Johan (2016-10-27). "Synchronous long-term oscillations in a synthetic gene circuit". Nature. 538 (7626): 514–517. Bibcode:2016Natur.538..514P. doi:10.1038/nature19841. PMC 5637407. PMID 27732583.
- Tyler, Jonathan; Shiu, Anne; Walton, Jay (2019-03-30). "Revisiting a synthetic intracellular regulatory network that exhibits oscillations". Journal of Mathematical Biology. 78 (7): 2341–2368. arXiv:1808.00595. doi:10.1007/s00285-019-01346-3. ISSN 1432-1416. PMID 30929046.
- Jolley, Craig C.; Ode, Koji L.; Ueda, Hiroki R. (2012). "A Design Principle for a Posttranslational Biochemical Oscillator". Cell Reports. 2 (4): 938–950. doi:10.1016/j.celrep.2012.09.006. ISSN 2211-1247. PMID 23084745.
- Garcia-Ojalvo, Jordi; Elowitz, Michael B.; Strogatz, Steven H. (2004-07-27). "Modeling a synthetic multicellular clock: Repressilators coupled by quorum sensing". Proceedings of the National Academy of Sciences of the United States of America. 101 (30): 10955–10960. Bibcode:2004PNAS..10110955G. doi:10.1073/pnas.0307095101. ISSN 0027-8424. PMC 503725. PMID 15256602.
- Gottlieb, Daphna (2019-06-29). "Agro-chronobiology: Integrating circadian clocks /time biology into storage management". Journal of Stored Products Research. 82: 9–16. doi:10.1016/j.jspr.2019.03.003. ISSN 0022-474X.
External links
- Direct link to the repressilator model and a Description in BioModels Database
- A simulation of the repressilator in R: https://gist.github.com/AndreyAkinshin/37f3e68a1576f9ea1e5c01f2fd64fe5e
- An online simulation of the repressilator: https://www.yschaerli.com/repressilator.html
- A diagram of the system of feedback loops in A. thaliana
- Direct link to information about the CCA1 gene and the role it plays in A. thaliana