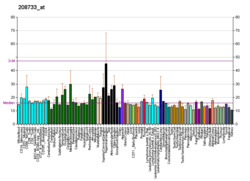
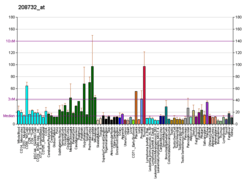
RAB2A
Ras-related protein Rab-2A is a protein that in humans is encoded by the RAB2A gene.[5]
Function
Members of the Rab protein family are nontransforming monomeric GTP-binding proteins of the Ras superfamily that contain 4 highly conserved regions involved in GTP binding and hydrolysis. Rabs are prenylated, membrane-bound proteins involved in vesicular fusion and trafficking. The mammalian RAB proteins show striking similarities to the S. cerevisiae YPT1 and SEC4 proteins, Ras-related GTP-binding proteins involved in the regulation of secretion.[supplied by OMIM][5]
Interactions
RAB2A has been shown to interact with GOLGA2.[6][7]
gollark: Most modern things will.
gollark: It's part of a more complex system, but basically:- Lasers (from Plethora) were on ComputerCraft turtles (robot things), which could fire them in arbitrary directions- The turtles ran a program which connected to a relay-type service I run on my web server, which let them receive commands like "fire at this position" or "fire in this direction"- That relay service passed commands from clients to turtles and the results back to said clients- The Python script connected to the MC server's dynmap (popular service for web maps for Minecraft servers) web API, which, among other things, provides positions of players, and sent commands to fire at the reported position of players.
gollark: Which aren't particularly big, but somewhat useful.
gollark: I have random Python scripts for things I wanted to do at some point which computers could do more easily than I could, like a̦̾̋p͍̫̿p͊̃̇l̜̋̓y̱ͫ̃i̴̔ͫn̲̲͡g͎͏̈́ ̯͋̿r̫͢͡a̲͜͝n̦̽̄d͈̮̤o̻̳̭ṃ̱ͦ ̼͌͠d̵̼̗ǐ̡̕ȧ̰̫ċ̔ͯr̀͠͠ì̄ͥt͓̼͌î͚̘c̞͋̀s͓̬̦ to text, controlling a bunch of laser turrets I had on a Minecraft server over the internet, bulk-converting some music to a different format, and generating beepy noises.
gollark: Putting together simple scripts or whatever to do some random task more easily.
References
- GRCh38: Ensembl release 89: ENSG00000104388 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000047187 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: RAB2A RAB2A, member RAS oncogene family".
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (Oct 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Short B, Preisinger C, Körner R, Kopajtich R, Byron O, Barr FA (Dec 2001). "A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic". The Journal of Cell Biology. 155 (6): 877–83. doi:10.1083/jcb.200108079. PMC 2150909. PMID 11739401.
Further reading
- Khosravi-Far R, Lutz RJ, Cox AD, Conroy L, Bourne JR, Sinensky M, Balch WE, Buss JE, Der CJ (Jul 1991). "Isoprenoid modification of rab proteins terminating in CC or CXC motifs". Proceedings of the National Academy of Sciences of the United States of America. 88 (14): 6264–8. doi:10.1073/pnas.88.14.6264. PMC 52063. PMID 1648736.
- Zahraoui A, Touchot N, Chardin P, Tavitian A (Jul 1989). "The human Rab genes encode a family of GTP-binding proteins related to yeast YPT1 and SEC4 products involved in secretion". The Journal of Biological Chemistry. 264 (21): 12394–401. PMID 2501306.
- Tachibana K, Umezawa A, Kato S, Takano T (Nov 1988). "Nucleotide sequence of a new YPT1-related human cDNA which belongs to the ras gene superfamily". Nucleic Acids Research. 16 (21): 10368. doi:10.1093/nar/16.21.10368. PMC 338870. PMID 3057444.
- Andersson B, Wentland MA, Ricafrente JY, Liu W, Gibbs RA (Apr 1996). "A "double adaptor" method for improved shotgun library construction". Analytical Biochemistry. 236 (1): 107–13. doi:10.1006/abio.1996.0138. PMID 8619474.
- Tisdale EJ, Balch WE (Nov 1996). "Rab2 is essential for the maturation of pre-Golgi intermediates". The Journal of Biological Chemistry. 271 (46): 29372–9. doi:10.1074/jbc.271.46.29372. PMID 8910601.
- Yu W, Andersson B, Worley KC, Muzny DM, Ding Y, Liu W, Ricafrente JY, Wentland MA, Lennon G, Gibbs RA (Apr 1997). "Large-scale concatenation cDNA sequencing". Genome Research. 7 (4): 353–8. doi:10.1101/gr.7.4.353. PMC 139146. PMID 9110174.
- de Leeuw HP, Koster PM, Calafat J, Janssen H, van Zonneveld AJ, van Mourik JA, Voorberg J (Oct 1998). "Small GTP-binding proteins in human endothelial cells". British Journal of Haematology. 103 (1): 15–9. doi:10.1046/j.1365-2141.1998.00965.x. PMID 9792283.
- Shisheva A, Chinni SR, DeMarco C (Sep 1999). "General role of GDP dissociation inhibitor 2 in membrane release of Rab proteins: modulations of its functional interactions by in vitro and in vivo structural modifications". Biochemistry. 38 (36): 11711–21. doi:10.1021/bi990200r. PMID 10512627.
- Caillol N, Pasqualini E, Lloubes R, Lombardo D (Sep 2000). "Impairment of bile salt-dependent lipase secretion in human pancreatic tumoral SOJ-6 cells". Journal of Cellular Biochemistry. 79 (4): 628–47. doi:10.1002/1097-4644(20001215)79:4<628::AID-JCB120>3.0.CO;2-T. PMID 10996854.
- Short B, Preisinger C, Körner R, Kopajtich R, Byron O, Barr FA (Dec 2001). "A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic". The Journal of Cell Biology. 155 (6): 877–83. doi:10.1083/jcb.200108079. PMC 2150909. PMID 11739401.
- Ni X, Ma Y, Cheng H, Jiang M, Guo L, Ji C, Gu S, Cao Y, Xie Y, Mao Y (2002). "Molecular cloning and characterization of a novel human Rab ( Rab2B) gene". Journal of Human Genetics. 47 (10): 548–51. doi:10.1007/s100380200083. PMID 12376746.
- Tisdale EJ (Dec 2003). "Rab2 interacts directly with atypical protein kinase C (aPKC) iota/lambda and inhibits aPKCiota/lambda-dependent glyceraldehyde-3-phosphate dehydrogenase phosphorylation". The Journal of Biological Chemistry. 278 (52): 52524–30. doi:10.1074/jbc.M309343200. PMID 14570876.
- Tisdale EJ, Kelly C, Artalejo CR (Dec 2004). "Glyceraldehyde-3-phosphate dehydrogenase interacts with Rab2 and plays an essential role in endoplasmic reticulum to Golgi transport exclusive of its glycolytic activity". The Journal of Biological Chemistry. 279 (52): 54046–52. doi:10.1074/jbc.M409472200. PMID 15485821.
- Chi A, Valencia JC, Hu ZZ, Watabe H, Yamaguchi H, Mangini NJ, Huang H, Canfield VA, Cheng KC, Yang F, Abe R, Yamagishi S, Shabanowitz J, Hearing VJ, Wu C, Appella E, Hunt DF (Nov 2006). "Proteomic and bioinformatic characterization of the biogenesis and function of melanosomes". Journal of Proteome Research. 5 (11): 3135–44. doi:10.1021/pr060363j. PMID 17081065.
- Dong C, Wu G (Nov 2007). "Regulation of anterograde transport of adrenergic and angiotensin II receptors by Rab2 and Rab6 GTPases". Cellular Signalling. 19 (11): 2388–99. doi:10.1016/j.cellsig.2007.07.017. PMC 2072516. PMID 17716866.
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.